Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile
- PMID: 33876817
- PMCID: PMC8212768
- DOI: 10.1093/jac/dkab097
Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile
Abstract
Background: Until recently, metronidazole was the first-line treatment for Clostridioides difficile infection and it is still commonly used. Though resistance has been reported due to the plasmid pCD-METRO, this does not explain all cases.
Objectives: To identify factors that contribute to plasmid-independent metronidazole resistance of C. difficile.
Methods: Here, we investigate resistance to metronidazole in a collection of clinical isolates of C. difficile using a combination of antimicrobial susceptibility testing on different solid agar media and WGS of selected isolates.
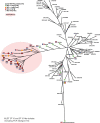
Results: We find that nearly all isolates demonstrate a haem-dependent increase in the MIC of metronidazole, which in some cases leads to isolates qualifying as resistant (MIC >2 mg/L). Moreover, we find an SNP in the haem-responsive gene hsmA, which defines a metronidazole-resistant lineage of PCR ribotype 010/MLST ST15 isolates that also includes pCD-METRO-containing strains.
Conclusions: Our data demonstrate that haem is crucial for medium-dependent metronidazole resistance in C. difficile.
© The Author(s) 2021. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures



References
-
- Chang JY, Antonopoulos DA, Kalra A. et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197: 435–8. - PubMed
-
- Rupnik M, Wilcox MH, Gerding DN.. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 2009; 7: 526–36. - PubMed
-
- Ooijevaar RE, van Beurden YH, Terveer EM. et al. Update of treatment algorithms for Clostridium difficile infection. Clin Microbiol Infect 2018; 24: 452–62. - PubMed
-
- McDonald LC, Gerding DN, Johnson S. et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66: 987–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

