Identification and characterization of repetitive DNA in the genus Didelphis Linnaeus, 1758 (Didelphimorphia, Didelphidae) and the use of satellite DNAs as phylogenetic markers
- PMID: 33877257
- PMCID: PMC8056902
- DOI: 10.1590/1678-4685-GMB-2020-0384
Identification and characterization of repetitive DNA in the genus Didelphis Linnaeus, 1758 (Didelphimorphia, Didelphidae) and the use of satellite DNAs as phylogenetic markers
Abstract
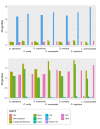
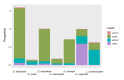
Didelphis species have been shown to exhibit very conservative karyotypes, which mainly differ in their constitutive heterochromatin, known to be mostly composed by repetitive DNAs. In this study, we used genome skimming data combined with computational pipelines to identify the most abundant repetitive DNA families of Lutreolina crassicaudata and all six Didelphis species. We found that transposable elements (TEs), particularly LINE-1, endogenous retroviruses, and SINEs, are the most abundant mobile elements in the studied species. Despite overall similar TE proportions, we report that species of the D. albiventris group consistently present a less diverse TE composition and smaller proportions of LINEs and LTRs in their genomes than other studied species. We also identified four new putative satDNAs (sat206, sat907, sat1430 and sat2324) in the genomes of Didelphis species, which show differences in abundance and nucleotide composition. Phylogenies based on satDNA sequences showed well supported relationships at the species (sat1430) and groups of species (sat206) level, recovering topologies congruent with previous studies. Our study is one of the first attempts to present a characterization of the most abundant families of repetitive DNAs of Lutreolina and Didelphis species providing insights into the repetitive DNA composition in the genome landscape of American marsupials.
Conflict of interest statement
Figures

Similar articles
-
Satellite DNAs-From Localized to Highly Dispersed Genome Components.Genes (Basel). 2023 Mar 17;14(3):742. doi: 10.3390/genes14030742. Genes (Basel). 2023. PMID: 36981013 Free PMC article. Review.
-
Heterochromatin evolution in Arachis investigated through genome-wide analysis of repetitive DNA.Planta. 2019 May;249(5):1405-1415. doi: 10.1007/s00425-019-03096-4. Epub 2019 Jan 24. Planta. 2019. PMID: 30680457
-
Structural and functional liaisons between transposable elements and satellite DNAs.Chromosome Res. 2015 Sep;23(3):583-96. doi: 10.1007/s10577-015-9483-7. Chromosome Res. 2015. PMID: 26293606 Review.
-
The Satellite DNAs Populating the Genome of Trigona hyalinata and the Sharing of a Highly Abundant satDNA in Trigona Genus.Genes (Basel). 2023 Feb 6;14(2):418. doi: 10.3390/genes14020418. Genes (Basel). 2023. PMID: 36833345 Free PMC article.
-
Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements.BMC Genomics. 2016 Dec 6;17(1):997. doi: 10.1186/s12864-016-3347-1. BMC Genomics. 2016. PMID: 27919246 Free PMC article.
Cited by
-
In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group.Genes (Basel). 2023 Jan 23;14(2):300. doi: 10.3390/genes14020300. Genes (Basel). 2023. PMID: 36833227 Free PMC article.
-
Satellite DNAs-From Localized to Highly Dispersed Genome Components.Genes (Basel). 2023 Mar 17;14(3):742. doi: 10.3390/genes14030742. Genes (Basel). 2023. PMID: 36981013 Free PMC article. Review.
References
-
- Amador LI, Giannini NP. Phylogeny and evolution of body mass in didelphid marsupials (Marsupialia: Didelphimorphia: Didelphidae) Org Divers Evol. 2016;16:641–657.
-
- Andrew S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. - PubMed
-
- Astúa D. Order Didelphimorphia. In: Wilson DE, Mittermeier RA, editors. Handbook of the Mammals of the World: Monotremes and Marsupials. Vol. 5. Lynx Edicions; Barcelona: 2015. 800
-
- Astúa D. Morphometrics of the largest new world marsupials, opossums of the genus Didelphis (Didelphimorphia, Didelphidae) Oecologia Aust. 2015;19:117–142.
LinkOut - more resources
Full Text Sources
Other Literature Sources

