Nutrient dominance governs the assembly of microbial communities in mixed nutrient environments
- PMID: 33877964
- PMCID: PMC8057819
- DOI: 10.7554/eLife.65948
Nutrient dominance governs the assembly of microbial communities in mixed nutrient environments
Abstract
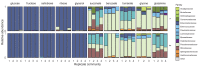
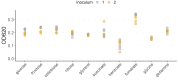
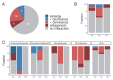
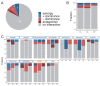
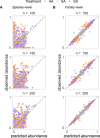
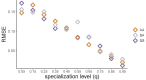
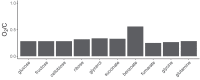
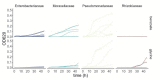
A major open question in microbial community ecology is whether we can predict how the components of a diet collectively determine the taxonomic composition of microbial communities. Motivated by this challenge, we investigate whether communities assembled in pairs of nutrients can be predicted from those assembled in every single nutrient alone. We find that although the null, naturally additive model generally predicts well the family-level community composition, there exist systematic deviations from the additive predictions that reflect generic patterns of nutrient dominance at the family level. Pairs of more-similar nutrients (e.g. two sugars) are on average more additive than pairs of more dissimilar nutrients (one sugar-one organic acid). Furthermore, sugar-acid communities are generally more similar to the sugar than the acid community, which may be explained by family-level asymmetries in nutrient benefits. Overall, our results suggest that regularities in how nutrients interact may help predict community responses to dietary changes.
Keywords: Microbial community assembly; computational biology; consumer-resource models; ecology; environmental complexity; nutrient interactions; resource specialization; systems biology.
© 2021, Estrela et al.
Conflict of interest statement
SE, AS, JV, AS No competing interests declared
Figures











References
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

