Substrate recruitment by zDHHC protein acyltransferases
- PMID: 33878949
- PMCID: PMC8059564
- DOI: 10.1098/rsob.210026
Substrate recruitment by zDHHC protein acyltransferases
Abstract
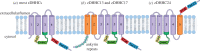
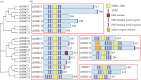
Protein palmitoylation is the post-translational attachment of fatty acids, most commonly palmitate (C16 : 0), onto a cysteine residue of a protein. This reaction is catalysed by a family of integral membrane proteins, the zDHHC protein acyltransferases (PATs), so-called due to the presence of an invariant Asp-His-His-Cys (DHHC) cysteine-rich domain harbouring the catalytic centre of the enzyme. Conserved throughout eukaryotes, the zDHHC PATs are encoded by multigene families and mediate palmitoylation of thousands of protein substrates. In humans, a number of zDHHC proteins are associated with human diseases, including intellectual disability, Huntington's disease, schizophrenia and cancer. Key to understanding the physiological and pathophysiological importance of individual zDHHC proteins is the identification of their protein substrates. Here, we will describe the approaches and challenges in assigning substrates for individual zDHHCs, highlighting key mechanisms that underlie substrate recruitment.
Keywords: fatty acylation; palmitoylation; post-translational modification; zDHHC enzyme.
Figures

References
-
- Broncel M, Serwa RA, Ciepla P, Krause E, Dallman MJ, Magee AI, Tate EW. 2015. Multifunctional reagents for quantitative proteome-wide analysis of protein modification in human cells and dynamic profiling of protein lipidation during vertebrate development. Angew. Chem. Int. Ed. 54, 5948. (10.1002/anie.201500342) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

