Analysis of miRNA-mediated regulation of flowering induction in Lilium × formolongi
- PMID: 33879043
- PMCID: PMC8058995
- DOI: 10.1186/s12870-021-02961-3
Analysis of miRNA-mediated regulation of flowering induction in Lilium × formolongi
Abstract
Background: MicroRNAs play pivotal roles in plant vegetative phase change and flowering induction via integrating into multiple flowering pathways. Lilium × formolongi is an important ornamental lily cultivar that can flower within one year after sowing. However, it remains unresolved how miRNA-mediated regulation networks contribute to the L. × formolongi characteristics of a short vegetative growth period and rapid flowering.
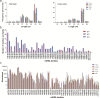
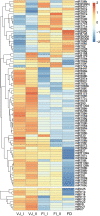
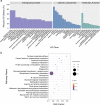
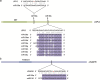
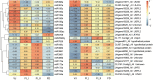
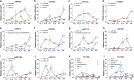
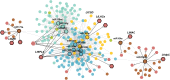
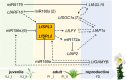
Results: In this study, the small RNA libraries and one degradome library were constructed for L. × formolongi during vegetative growth and flowering initiation, and 366 conserved miRNAs and 32 novel miRNAs were identified. Additionally, 84 miRNAs were significantly differentially expressed during development. A total of 396 targets of 185 miRNAs were identified and validated through degradome sequencing. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses showed that functions of the targets were top enriched in the cold and cadmium ion responses, pentose phosphate pathway and carbon fixation in photosynthetic organisms. Furthermore, among 23 differentially expressed miRNA-target pairs, the miR156s-LfSPL2, miR172a-LfAP2 and miR164a-LfNAC pairs as well as miR159a-LfSPL2 were found to be relevant to flowering based on the correlation analysis of expression profiles in the miRNA libraries, degradome and transcriptome. A coexpression regulatory network focused on differentially expressed pairs was also constructed by WGCNA, and 14 miRNAs were considered putative key miRNAs during vegetative development and flowering induction. miR156a/ d/ e showed particularly strong relationships with other miRNAs in the coexpression network.
Conclusions: This study provides cues for the further exploration of the regulatory mechanisms of short vegetative development and flowering in L. × formolongi.
Keywords: Development; Flowering; Lilium × formolongi; miRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Analysis of global gene expression profiles during the flowering initiation process of Lilium × formolongi.Plant Mol Biol. 2017 Jul;94(4-5):361-379. doi: 10.1007/s11103-017-0612-x. Epub 2017 Apr 20. Plant Mol Biol. 2017. PMID: 28429252
-
Identification of trehalose-6-phosphate synthase (TPS)-coding genes involved in flowering induction of Lilium× formolongi.Plant Physiol Biochem. 2022 Jan 15;171:84-94. doi: 10.1016/j.plaphy.2021.12.025. Epub 2021 Dec 27. Plant Physiol Biochem. 2022. PMID: 34973503
-
Cryptochrome 2 from Lilium × formolongi Regulates Photoperiodic Flowering in Transgenic Arabidopsis thaliana.Int J Mol Sci. 2021 Nov 29;22(23):12929. doi: 10.3390/ijms222312929. Int J Mol Sci. 2021. PMID: 34884732 Free PMC article.
-
To bloom or not to bloom: role of microRNAs in plant flowering.Mol Plant. 2015 Mar;8(3):359-77. doi: 10.1016/j.molp.2014.12.018. Epub 2014 Dec 31. Mol Plant. 2015. PMID: 25737467 Review.
-
Small but powerful: function of microRNAs in plant development.Plant Cell Rep. 2018 Mar;37(3):515-528. doi: 10.1007/s00299-017-2246-5. Epub 2018 Jan 9. Plant Cell Rep. 2018. PMID: 29318384 Review.
Cited by
-
Screening and validation of optimal miRNA reference genes in different developing stages and tissues of Lilium henryi Baker.Sci Rep. 2024 Jan 17;14(1):1545. doi: 10.1038/s41598-024-51562-1. Sci Rep. 2024. PMID: 38233457 Free PMC article.
-
The Establishment of a Genetic Transformation System and the Acquisition of Transgenic Plants of Oriental Hybrid Lily (Lilium L.).Int J Mol Sci. 2023 Jan 2;24(1):782. doi: 10.3390/ijms24010782. Int J Mol Sci. 2023. PMID: 36614225 Free PMC article.
-
Integrated analysis of miRNAome transcriptome and degradome reveals miRNA-target modules governing floral florescence development and senescence across early- and late-flowering genotypes in tree peony.Front Plant Sci. 2022 Dec 14;13:1082415. doi: 10.3389/fpls.2022.1082415. eCollection 2022. Front Plant Sci. 2022. PMID: 36589111 Free PMC article.
-
The Pivotal Role of Noncoding RNAs in Flowering Time Regulation.Genes (Basel). 2023 Nov 23;14(12):2114. doi: 10.3390/genes14122114. Genes (Basel). 2023. PMID: 38136936 Free PMC article. Review.
References
-
- Fortanier EJ. Reviewing the length of the generation period and its shortening, particularly in tulips. Sci Hort. 1973;1(1):107–116. doi: 10.1016/0304-4238(73)90010-1. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

