A flexible ChIP-sequencing simulation toolkit
- PMID: 33879052
- PMCID: PMC8056602
- DOI: 10.1186/s12859-021-04097-5
A flexible ChIP-sequencing simulation toolkit
Abstract
Background: A major challenge in evaluating quantitative ChIP-seq analyses, such as peak calling and differential binding, is a lack of reliable ground truth data. Accurate simulation of ChIP-seq data can mitigate this challenge, but existing frameworks are either too cumbersome to apply genome-wide or unable to model a number of important experimental conditions in ChIP-seq.
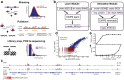
Results: We present ChIPs, a toolkit for rapidly simulating ChIP-seq data using statistical models of key experimental steps. We demonstrate how ChIPs can be used for a range of applications, including benchmarking analysis tools and evaluating the impact of various experimental parameters. ChIPs is implemented as a standalone command-line program written in C++ and is available from https://github.com/gymreklab/chips .
Conclusions: ChIPs is an efficient ChIP-seq simulation framework that generates realistic datasets over a flexible range of experimental conditions. It can serve as an important component in various ChIP-seq analyses where ground truth data are needed.
Keywords: Bioinformatics; ChIP-sequencing; Command-line program; Epigenomics; Simulation tool.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin SF, Palmieri C, Caldas C, Carroll JS. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

