A pocket-factor-triggered conformational switch in the hepatitis B virus capsid
- PMID: 33879615
- PMCID: PMC8092406
- DOI: 10.1073/pnas.2022464118
A pocket-factor-triggered conformational switch in the hepatitis B virus capsid
Abstract
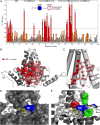
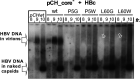
Viral hepatitis is growing into an epidemic illness, and it is urgent to neutralize the main culprit, hepatitis B virus (HBV), a small-enveloped retrotranscribing DNA virus. An intriguing observation in HB virion morphogenesis is that capsids with immature genomes are rarely enveloped and secreted. This prompted, in 1982, the postulate that a regulated conformation switch in the capsid triggers envelopment. Using solid-state NMR, we identified a stable alternative conformation of the capsid. The structural variations focus on the hydrophobic pocket of the core protein, a hot spot in capsid-envelope interactions. This structural switch is triggered by specific, high-affinity binding of a pocket factor. The conformational change induced by the binding is reminiscent of a maturation signal. This leads us to formulate the "synergistic double interaction" hypothesis, which explains the regulation of capsid envelopment and indicates a concept for therapeutic interference with HBV envelopment.
Keywords: Triton; hepatitis B virus; hydrophobic pocket; solid-state NMR.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Graber-Stiehl I., The silent epidemic killing more people than HIV, malaria or TB. Nature 564, 24–26 (2018). - PubMed
-
- Nassal M., HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984 (2015). - PubMed
-
- Seitz S., Habjanič J., Schütz A. K., Bartenschlager R., The hepatitis B virus envelope proteins: Molecular gymnastics throughout the viral life cycle. Annu. Rev. Virol. 7, 263–288 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

