The Degenerate Tale of Ascidian Tails
- PMID: 33881514
- PMCID: PMC10452958
- DOI: 10.1093/icb/icab022
The Degenerate Tale of Ascidian Tails
Abstract
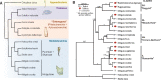
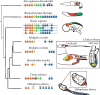
Ascidians are invertebrate chordates, with swimming chordate tadpole larvae that have distinct heads and tails. The head contains the small brain, sensory organs, including the ocellus (light) and otolith (gravity) and the presumptive endoderm, while the tail has a notochord surrounded by muscle cells and a dorsal nerve cord. One of the chordate features is a post-anal tail. Ascidian tadpoles are nonfeeding, and their tails are critical for larval locomotion. After hatching the larvae swim up toward light and are carried by the tide and ocean currents. When competent to settle, ascidian tadpole larvae swim down, away from light, to settle and metamorphose into a sessile adult. Tunicates are classified as chordates because of their chordate tadpole larvae; in contrast, the sessile adult has a U-shaped gut and very derived body plan, looking nothing like a chordate. There is one group of ascidians, the Molgulidae, where many species are known to have tailless larvae. The Swalla Lab has been studying the evolution of tailless ascidian larvae in this clade for over 30 years and has shown that tailless larvae have evolved independently several times in this clade. Comparison of the genomes of two closely related species, the tailed Molgula oculata and tailless Molgula occulta reveals much synteny, but there have been multiple insertions and deletions that have disrupted larval genes in the tailless species. Genomics and transcriptomics have previously shown that there are pseudogenes expressed in the tailless embryos, suggesting that the partial rescue of tailed features in their hybrid larvae is due to the expression of intact genes from the tailed parent. Yet surprisingly, we find that the notochord gene regulatory network is mostly intact in the tailless M. occulta, although the notochord does not converge and extend and remains as an aggregate of cells we call the "notoball." We expect that eventually many of the larval gene networks will become evolutionarily lost in tailless ascidians and the larval body plan abandoned, with eggs developing directly into an adult. Here we review the current evolutionary and developmental evidence on how the molgulids lost their tails.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Integrative and Comparative Biology.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures
References
-
- Ambrosino L, Vassalli QA, D’Agostino Y, Esposito R, Cetrangolo V, Caputi L, Locascio A, Aniello F, D’Aniello S, Chatzigeorgiou M, et al. 2019. Functional conserved non-coding elements among tunicates and chordates. Dev Biol 448:101–10. - PubMed
-
- Amemiya CT, Prohaska SJ, Hill-Force A, Cook A, Wasserscheid J, Ferrier DEK, Pascual-Anaya J, Garcia-Ferna`ndez J, Dewar K, Stadler PF. 2008. The amphioxus Hox cluster: characterization, comparative genomics, and evolution. J Exp Zool 310:465–77. - PubMed
-
- Aronowicz J, Lowe C. 2006. Hox gene expression in the hemichordate Saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Int Comp Biol 46:890–901. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

