Evolution of pathogen response genes associated with increased disease susceptibility during adaptation to an extreme drought in a Brassica rapa plant population
- PMID: 33882815
- PMCID: PMC8060997
- DOI: 10.1186/s12862-021-01789-7
Evolution of pathogen response genes associated with increased disease susceptibility during adaptation to an extreme drought in a Brassica rapa plant population
Abstract
Background: Pathogens are key components in natural and agricultural plant systems. There is evidence of evolutionary changes in disease susceptibility as a consequence of climate change, but we know little about the underlying genetic basis of this evolution. To address this, we took advantage of a historical seed collection of a Brassica rapa population, which we previously demonstrated evolved an increase in disease susceptibility to a necrotrophic fungal pathogen following a drought.
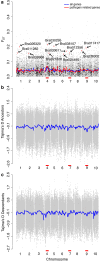
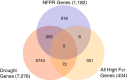
Results: Previously, we combined a resurrection experiment with genome-wide sequencing of 124 pooled ancestral and descendant plants. Here, using these previously generated sequence data (Franks et al. in Mol Ecol 25(15):3622-3631, 2016), we show that well-characterized necrotrophic fungal pathogen response (NFPR) genes have evolved, as indicated by changes in allele frequency, between ancestors and descendants, with several of them identified as extreme FST outliers. The jasmonic acid (JA) signaling pathway in particular seems to underlie the evolution of disease susceptibility, in addition to its well characterized role in plastic disease response. We identify a list of 260 genes that are both NFPR genes and are differentially expressed in response to drought, based on publicly available data. We present evidence that five of these genes evolved between ancestors and descendants, suggesting that the drought acted as the evolutionary driver, and that the accompanying increase in disease susceptibility may have been a consequence of genetic pleiotropy.
Conclusions: Our study provides evidence that for this population, standing variation in NFPR genes is affected by natural selection related to climate change. Our results reveal potentially important candidates that may underlie trait evolution in both crops and natural systems. Additionally, this trade-off between adaptation to biotic and abiotic stresses is an example of how climate change can have diverse and unexpected consequences.
Keywords: Alternaria brassicae; Brassica rapa; Climate change; Drought; Necrotrophic fungal pathogen response; Resurrection approach.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

References
-
- Desprez-Loustau ML, Marcais B, Nageleisen L, Piou D, Vannini A. Interactive effects of drought and pathogens in forest trees. Ann For Sci. 2006;63:597–612. doi: 10.1051/forest:2006040. - DOI
-
- Garrett K, Thomas-Sharma S, Forbes G, Nopsa J. Climate change and plant pathogen invasions. In: Ziska L, Dukes J, editors. Invasive species and global climate change. Wallingford: CABI; 2014.
-
- Pautasso M, Doring T, Garbelotto M, Lorenzo P, Jeger M. Impacts of climate change on plant disease—opinions and trends. Eur J Plant Pathol. 2012;133:295–313. doi: 10.1007/s10658-012-9936-1. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
