IFN-λ4 genetic variants influence clinical malaria episodes in a cohort of Kenyan children
- PMID: 33882912
- PMCID: PMC8058600
- DOI: 10.1186/s12936-021-03689-z
IFN-λ4 genetic variants influence clinical malaria episodes in a cohort of Kenyan children
Abstract
Background: Interferon (IFN)- λ4, a type III IFN, production is controlled by a dinucleotide frameshift variant (rs368234815-dG/TT) within the first exon of the IFNL4 gene. Carriers of the IFNL4-dG allele but not the IFNL4-TT allele are able to produce the IFN-λ4 protein. Patients with hepatitis C virus that do not produce the IFN-λ4 protein have higher rates of viral clearance suggesting a potential inhibitory role of IFN-λ4 in liver-tropic infections.
Methods: In this study, it was investigated whether children infected with Plasmodium falciparum, which has a well-characterized liver stage infection, would be more susceptible to clinical malaria relative to their IFNL4-rs368234815 allele. A cohort of 122 children from a malaria holoendemic region of Kenya was analysed. Episodes of clinical malaria and upper respiratory tract infections (URTIs) were determined using information collected from birth to 2 years of age. The dinucleotide frameshift variant IFNL4-rs368234815-dG/TT was genotyped using a TaqMan assay.
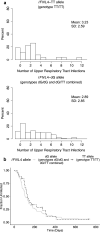
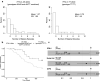
Results: In this cohort, 33% of the study participants had the dG/dG genotype, 45% had the dG/TT genotype, and 22% had TT/TT genotype. The number and time to first episode of clinical malaria and URTIs with respect to the IFNL4-rs368234815 allele was evaluated. It was found that children that carried the IFNL4-rs368234815-dG allele had an increased number of clinical malaria episodes. In addition, there was a significant association between earlier age of first malaria infection with carriers of the IFNL4-dG allele (p-value: 0.021).
Conclusion: The results suggest that the ability to produce IFN-λ4 negatively affects host immune protection against P. falciparum malaria in Kenyan children.
Keywords: IFN-λ4; Innate Immunity; Malaria; Plasmodium falciparum; Type III Interferon.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
IFNL4 Genotypes and Risk of Childhood Burkitt Lymphoma in East Africa.J Interferon Cytokine Res. 2023 Sep;43(9):394-402. doi: 10.1089/jir.2023.0014. Epub 2023 Jun 27. J Interferon Cytokine Res. 2023. PMID: 37366802 Free PMC article.
-
IFN-λ4 is associated with increased risk and earlier occurrence of several common infections in African children.Genes Immun. 2021 May;22(1):44-55. doi: 10.1038/s41435-021-00127-7. Epub 2021 Apr 13. Genes Immun. 2021. PMID: 33850301 Free PMC article.
-
A Novel Inactive Isoform with a Restored Reading Frame Is Expressed from the Human Interferon Lambda 4 TT Allele at rs368234815.J Interferon Cytokine Res. 2023 Sep;43(9):370-378. doi: 10.1089/jir.2022.0199. Epub 2023 Mar 7. J Interferon Cytokine Res. 2023. PMID: 36880961 Free PMC article.
-
What Have We Learned from Studies of IFN-λ Variants and Hepatitis C Virus Infection?J Interferon Cytokine Res. 2019 Oct;39(10):618-626. doi: 10.1089/jir.2019.0048. Epub 2019 Jun 4. J Interferon Cytokine Res. 2019. PMID: 31161939 Free PMC article. Review.
-
Identifying causal variants at the interferon lambda locus in case-control studies: Utilizing non-synonymous variant rs117648444 to probe the role of IFN-λ4.Gene. 2018 Jul 20;664:168-180. doi: 10.1016/j.gene.2018.04.076. Epub 2018 Apr 27. Gene. 2018. PMID: 29705128 Review.
Cited by
-
IFNL4 Genotypes and Risk of Childhood Burkitt Lymphoma in East Africa.J Interferon Cytokine Res. 2023 Sep;43(9):394-402. doi: 10.1089/jir.2023.0014. Epub 2023 Jun 27. J Interferon Cytokine Res. 2023. PMID: 37366802 Free PMC article.
-
Polymorphic pseudogenes in the human genome - a comprehensive assessment.Hum Genet. 2024 Dec;143(12):1465-1479. doi: 10.1007/s00439-024-02715-9. Epub 2024 Nov 2. Hum Genet. 2024. PMID: 39488654 Free PMC article.
-
Distinct molecular phenotypes involving several human diseases are induced by IFN-λ3 and IFN-λ4 in monocyte-derived macrophages.Genes Immun. 2022 Apr;23(2):73-84. doi: 10.1038/s41435-022-00164-w. Epub 2022 Feb 3. Genes Immun. 2022. PMID: 35115664 Free PMC article.
-
The Immunomodulatory Role of Estrogen in Malaria: A Review of Sex Differences and Therapeutic Implications.Immun Inflamm Dis. 2025 Feb;13(2):e70148. doi: 10.1002/iid3.70148. Immun Inflamm Dis. 2025. PMID: 39898752 Free PMC article. Review.
References
-
- WHO. World malaria report . 20 years of global progress and challenges. Geneva: World Health Organization; 2020. p. 2020.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

