Single-Dilution COVID-19 Antibody Test with Qualitative and Quantitative Readouts
- PMID: 33883259
- PMCID: PMC8546701
- DOI: 10.1128/mSphere.00224-21
Single-Dilution COVID-19 Antibody Test with Qualitative and Quantitative Readouts
Abstract
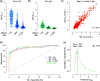
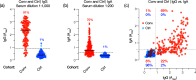
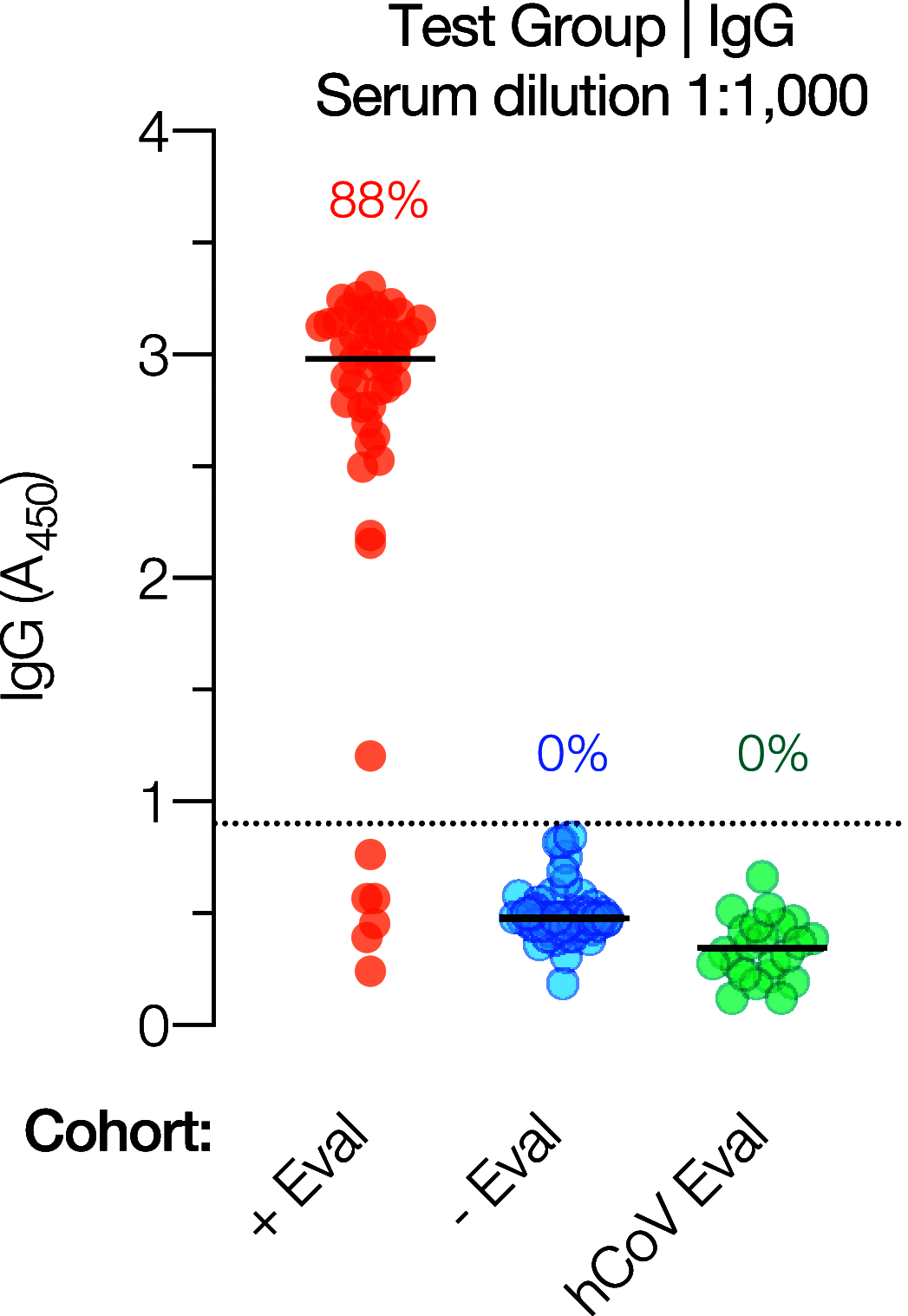
The coronavirus disease 2019 (COVID-19) global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to place an immense burden on societies and health care systems. A key component of COVID-19 control efforts is serological testing to determine the community prevalence of SARS-CoV-2 exposure and quantify individual immune responses to prior SARS-CoV-2 infection or vaccination. Here, we describe a laboratory-developed antibody test that uses readily available research-grade reagents to detect SARS-CoV-2 exposure in patient blood samples with high sensitivity and specificity. We further show that this sensitive test affords the estimation of viral spike-specific IgG titers from a single sample measurement, thereby providing a simple and scalable method to measure the strength of an individual's immune response. The accuracy, adaptability, and cost-effectiveness of this test make it an excellent option for clinical deployment in the ongoing COVID-19 pandemic.IMPORTANCE Serological surveillance has become an important public health tool during the COVID-19 pandemic. Detection of protective antibodies and seroconversion after SARS-CoV-2 infection or vaccination can help guide patient care plans and public health policies. Serology tests can detect antibodies against past infections; consequently, they can help overcome the shortcomings of molecular tests, which can detect only active infections. This is important, especially when considering that many COVID-19 patients are asymptomatic. In this study, we describe an enzyme-linked immunosorbent assay (ELISA)-based qualitative and quantitative serology test developed to measure IgG and IgA antibodies against the SARS-CoV-2 spike glycoprotein. The test can be deployed using commonly available laboratory reagents and equipment and displays high specificity and sensitivity. Furthermore, we demonstrate that IgG titers in patient samples can be estimated from a single measurement, enabling the assay's use in high-throughput clinical environments.
Keywords: COVID-19; IgA; IgG; SARS-CoV-2; laboratory diagnostic test; quantitative test; serology; spike protein.
Copyright © 2021 Bortz et al.
Figures




Update of
-
Development, clinical translation, and utility of a COVID-19 antibody test with qualitative and quantitative readouts.medRxiv [Preprint]. 2020 Sep 11:2020.09.10.20192187. doi: 10.1101/2020.09.10.20192187. medRxiv. 2020. Update in: mSphere. 2021 Apr 21;6(2):e00224-21. doi: 10.1128/mSphere.00224-21. PMID: 32935116 Free PMC article. Updated. Preprint.
References
-
- Johns Hopkins University. 23 February 2021, accession date. Johns Hopkins University of Medicine coronavirus resource center. Johns Hopkins University, Baltimore, MD. https://coronavirus.jhu.edu/.
Publication types
MeSH terms
Substances
Grants and funding
- AI13261/AI/NIAID NIH HHS/United States
- R01 AI124753/AI/NIAID NIH HHS/United States
- K08 CA143153/CA/NCI NIH HHS/United States
- R01 AI134753/AI/NIAID NIH HHS/United States
- R01 CA236546/CA/NCI NIH HHS/United States
- T32 GM007288/GM/NIGMS NIH HHS/United States
- R01 AI145024/AI/NIAID NIH HHS/United States
- R01 AI132633/AI/NIAID NIH HHS/United States
- F30 AI150055/AI/NIAID NIH HHS/United States
- R01 AI125462/AI/NIAID NIH HHS/United States
- 2T32GM007288-45/GM/NIGMS NIH HHS/United States
- P30 AI124414/AI/NIAID NIH HHS/United States
- UL1 TR002556/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
