HIM-17 regulates the position of recombination events and GSP-1/2 localization to establish short arm identity on bivalents in meiosis
- PMID: 33883277
- PMCID: PMC8092412
- DOI: 10.1073/pnas.2016363118
HIM-17 regulates the position of recombination events and GSP-1/2 localization to establish short arm identity on bivalents in meiosis
Abstract
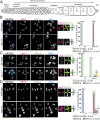
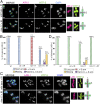
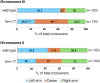
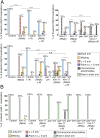
The position of recombination events established along chromosomes in early prophase I and the chromosome remodeling that takes place in late prophase I are intrinsically linked steps of meiosis that need to be tightly regulated to ensure accurate chromosome segregation and haploid gamete formation. Here, we show that RAD-51 foci, which form at the sites of programmed meiotic DNA double-strand breaks (DSBs), exhibit a biased distribution toward off-centered positions along the chromosomes in wild-type Caenorhabditis elegans, and we identify two meiotic roles for chromatin-associated protein HIM-17 that ensure normal chromosome remodeling in late prophase I. During early prophase I, HIM-17 regulates the distribution of DSB-dependent RAD-51 foci and crossovers on chromosomes, which is critical for the formation of distinct chromosome subdomains (short and long arms of the bivalents) later during chromosome remodeling. During late prophase I, HIM-17 promotes the normal expression and localization of protein phosphatases GSP-1/2 to the surface of the bivalent chromosomes and may promote GSP-1 phosphorylation, thereby antagonizing Aurora B kinase AIR-2 loading on the long arms and preventing premature loss of sister chromatid cohesion. We propose that HIM-17 plays distinct roles at different stages during meiotic progression that converge to promote normal chromosome remodeling and accurate chromosome segregation.
Keywords: DNA double-strand breaks; HIM-17; crossovers; late prophase I remodeling; meiosis.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Caenorhabditis elegans RMI2 functional homolog-2 (RMIF-2) and RMI1 (RMH-1) have both overlapping and distinct meiotic functions within the BTR complex.PLoS Genet. 2021 Jul 12;17(7):e1009663. doi: 10.1371/journal.pgen.1009663. eCollection 2021 Jul. PLoS Genet. 2021. PMID: 34252074 Free PMC article.
-
The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance.PLoS Genet. 2013;9(8):e1003674. doi: 10.1371/journal.pgen.1003674. Epub 2013 Aug 8. PLoS Genet. 2013. PMID: 23950729 Free PMC article.
-
Chromatin landscape, DSB levels, and cKU-70/80 contribute to patterning of meiotic DSB processing along chromosomes in C. elegans.PLoS Genet. 2023 Jan 27;19(1):e1010627. doi: 10.1371/journal.pgen.1010627. eCollection 2023 Jan. PLoS Genet. 2023. PMID: 36706157 Free PMC article.
-
Meiotic recombination and the crossover assurance checkpoint in Caenorhabditis elegans.Semin Cell Dev Biol. 2016 Jun;54:106-16. doi: 10.1016/j.semcdb.2016.03.014. Epub 2016 Mar 21. Semin Cell Dev Biol. 2016. PMID: 27013114 Free PMC article. Review.
-
Distribution of meiotic recombination events: talking to your neighbors.Curr Opin Genet Dev. 2009 Apr;19(2):105-12. doi: 10.1016/j.gde.2009.02.005. Epub 2009 Mar 26. Curr Opin Genet Dev. 2009. PMID: 19328674 Free PMC article. Review.
Cited by
-
DNA repair, recombination, and damage signaling.Genetics. 2022 Feb 4;220(2):iyab178. doi: 10.1093/genetics/iyab178. Genetics. 2022. PMID: 35137093 Free PMC article. Review.
-
Overlapping and separable activities of BRA-2 and HIM-17 promote occurrence and regulation of pairing and synapsis during Caenorhabditis elegans meiosis.Nat Commun. 2025 Mar 13;16(1):2516. doi: 10.1038/s41467-025-57862-y. Nat Commun. 2025. PMID: 40082424 Free PMC article.
-
The conserved ATPase PCH-2 controls the number and distribution of crossovers by antagonizing their formation in Caenorhabditis elegans.Elife. 2025 Feb 18;13:RP102409. doi: 10.7554/eLife.102409. Elife. 2025. PMID: 39964851 Free PMC article.
-
Widespread transposon co-option in the Caenorhabditis germline regulatory network.Sci Adv. 2022 Dec 16;8(50):eabo4082. doi: 10.1126/sciadv.abo4082. Epub 2022 Dec 16. Sci Adv. 2022. PMID: 36525485 Free PMC article.
-
PCH-2 and meiotic HORMADs: A module for evolutionary innovation in meiosis?Curr Top Dev Biol. 2023;151:317-344. doi: 10.1016/bs.ctdb.2022.07.001. Epub 2022 Jul 28. Curr Top Dev Biol. 2023. PMID: 36681475 Free PMC article. Review.
References
-
- Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). - PubMed
-
- Bergerat A., et al. ., An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417 (1997). - PubMed
-
- Dernburg A. F., et al. ., Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

