GLUT1 overexpression enhances glucose metabolism and promotes neonatal heart regeneration
- PMID: 33883682
- PMCID: PMC8060418
- DOI: 10.1038/s41598-021-88159-x
GLUT1 overexpression enhances glucose metabolism and promotes neonatal heart regeneration
Abstract
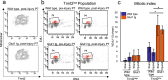
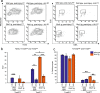
The mammalian heart switches its main metabolic substrate from glucose to fatty acids shortly after birth. This metabolic switch coincides with the loss of regenerative capacity in the heart. However, it is unknown whether glucose metabolism regulates heart regeneration. Here, we report that glucose metabolism is a determinant of regenerative capacity in the neonatal mammalian heart. Cardiac-specific overexpression of Glut1, the embryonic form of constitutively active glucose transporter, resulted in an increase in glucose uptake and concomitant accumulation of glycogen storage in postnatal heart. Upon cryoinjury, Glut1 transgenic hearts showed higher regenerative capacity with less fibrosis than non-transgenic control hearts. Interestingly, flow cytometry analysis revealed two distinct populations of ventricular cardiomyocytes: Tnnt2-high and Tnnt2-low cardiomyocytes, the latter of which showed significantly higher mitotic activity in response to high intracellular glucose in Glut1 transgenic hearts. Metabolic profiling shows that Glut1-transgenic hearts have a significant increase in the glucose metabolites including nucleotides upon injury. Inhibition of the nucleotide biosynthesis abrogated the regenerative advantage of high intra-cardiomyocyte glucose level, suggesting that the glucose enhances the cardiomyocyte regeneration through the supply of nucleotides. Our data suggest that the increase in glucose metabolism promotes cardiac regeneration in neonatal mouse heart.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

