Post-transcriptional control of T-cell cytokine production: Implications for cancer therapy
- PMID: 33884612
- PMCID: PMC8358718
- DOI: 10.1111/imm.13339
Post-transcriptional control of T-cell cytokine production: Implications for cancer therapy
Abstract
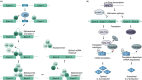
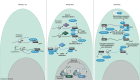
As part of the adaptive immune system, T cells are vital for the eradication of infected and malignantly transformed cells. To perform their protective function, T cells produce effector molecules that are either directly cytotoxic, such as granzymes, perforin, interferon-γ and tumour necrosis factor α, or attract and stimulate (immune) cells, such as interleukin-2. As these molecules can also induce immunopathology, tight control of their production is required. Indeed, inflammatory cytokine production is regulated on multiple levels. Firstly, locus accessibility and transcription factor availability and activity determine the amount of mRNA produced. Secondly, post-transcriptional mechanisms, influencing mRNA splicing/codon usage, stability, decay, localization and translation rate subsequently determine the amount of protein that is produced. In the immune suppressive environments of tumours, T cells gradually lose the capacity to produce effector molecules, resulting in tumour immune escape. Recently, the role of post-transcriptional regulation in fine-tuning T-cell effector function has become more appreciated. Furthermore, several groups have shown that exhausted or dysfunctional T cells from cancer patients or murine models possess mRNA for inflammatory mediators, but fail to produce effector molecules, hinting that post-transcriptional events also play a role in hampering tumour-infiltrating lymphocyte effector function. Here, the post-transcriptional regulatory events governing T-cell cytokine production are reviewed, with a specific focus on the importance of post-transcriptional regulation in anti-tumour responses. Furthermore, potential approaches to circumvent tumour-mediated dampening of T-cell effector function through the (dis)engagement of post-transcriptional events are explored, such as CRISPR/Cas9-mediated genome editing or chimeric antigen receptors.
Keywords: AU-rich elements; T cells; cancer; effector function; post-transcriptional regulation.
© 2021 John Wiley & Sons Ltd.
Conflict of interest statement
The author has no competing interests to declare.
Figures

Similar articles
-
Employing CRISPR-Cas9 to Enhance T Cell Effector Function.Methods Mol Biol. 2024;2782:195-208. doi: 10.1007/978-1-0716-3754-8_16. Methods Mol Biol. 2024. PMID: 38622404
-
Using CRISPR to enhance T cell effector function for therapeutic applications.Cytokine X. 2020 Dec 21;3(1):100049. doi: 10.1016/j.cytox.2020.100049. eCollection 2021 Mar. Cytokine X. 2020. PMID: 33604565 Free PMC article. Review.
-
Toll-like receptor-2/7-mediated T cell activation: An innate potential to augment CD8+ T cell cytokine production.Scand J Immunol. 2021 May;93(5):e13019. doi: 10.1111/sji.13019. Epub 2021 Jan 19. Scand J Immunol. 2021. PMID: 33377182 Review.
-
Renal Cell Carcinoma-Infiltrating CD3low Vγ9Vδ1 T Cells Represent Potentially Novel Anti-Tumor Immune Players.Curr Issues Mol Biol. 2021 May 27;43(1):226-239. doi: 10.3390/cimb43010019. Curr Issues Mol Biol. 2021. PMID: 34071865 Free PMC article.
-
CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review.J Cell Physiol. 2019 Jun;234(6):8509-8521. doi: 10.1002/jcp.27782. Epub 2018 Nov 22. J Cell Physiol. 2019. PMID: 30520029 Review.
Cited by
-
Interaction between gut microbiota and immune checkpoint inhibitor-related colitis.Front Immunol. 2022 Oct 27;13:1001623. doi: 10.3389/fimmu.2022.1001623. eCollection 2022. Front Immunol. 2022. PMID: 36389768 Free PMC article. Review.
-
Multiple roles for AU-rich RNA binding proteins in the development of haematologic malignancies and their resistance to chemotherapy.RNA Biol. 2024 Jan;21(1):1-17. doi: 10.1080/15476286.2024.2346688. Epub 2024 May 27. RNA Biol. 2024. PMID: 38798162 Free PMC article. Review.
-
Targeting a disintegrin and metalloprotease (ADAM) 17-CD122 axis enhances CD8+ T cell effector differentiation and anti-tumor immunity.Signal Transduct Target Ther. 2024 Jun 26;9(1):152. doi: 10.1038/s41392-024-01873-6. Signal Transduct Target Ther. 2024. PMID: 38918390 Free PMC article.
-
Circ-CAMTA1 regulated by Ca2+ influx inhibited pyruvate carboxylase activity and modulate T cell function in patients with systemic lupus erythematosus.Arthritis Res Ther. 2024 Oct 29;26(1):185. doi: 10.1186/s13075-024-03422-6. Arthritis Res Ther. 2024. PMID: 39473004 Free PMC article.
-
Employing CRISPR-Cas9 to Enhance T Cell Effector Function.Methods Mol Biol. 2024;2782:195-208. doi: 10.1007/978-1-0716-3754-8_16. Methods Mol Biol. 2024. PMID: 38622404
References
-
- Bisping G, Lugering N, Lutke‐Brintrup S, Pauels HG, Schurmann G, Domschke W, et al. Patients with inflammatory bowel disease (IBD) reveal increased induction capacity of intracellular interferon‐gamma (IFN‐g) in peripheral CD8+ lymphocytes co‐cultured with intestinal epithelial cells. Clin Exp Immunol. 2001;123:15–22. - PMC - PubMed
-
- Rafa H, Amri M, Saoula H, Belkhelfa M, Medjeber O, Boutaleb A. Involvement of Interferon‐γ in bowel disease pathogenesis by nitric oxide pathway: A study in algerian patients. J Interf Cytokine Res. 2010;30:691–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

