Impact of splicing mutations in acute myeloid leukemia treated with hypomethylating agents combined with venetoclax
- PMID: 33885753
- PMCID: PMC8095152
- DOI: 10.1182/bloodadvances.2020004173
Impact of splicing mutations in acute myeloid leukemia treated with hypomethylating agents combined with venetoclax
Abstract
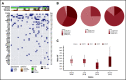
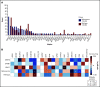
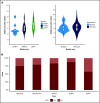
Spliceosome mutations (SRSF2, SF3B1, U2AF1, ZRSR2), are encountered in ∼50% of secondary acute myeloid leukemia cases (sAML) and define a molecular subgroup with outcomes similar to sAML in de novo AML patients treated with intensive chemotherapy. Outcomes in patients with spliceosome mutations treated with hypomethylating agents in combination with venetoclax (HMA+VEN) remains unknown. The primary objective was to compare outcomes in patients with spliceosome mutations vs wild-type patients treated with HMA+VEN. Secondary objectives included analysis of the mutational landscape of the spliceosome cohort and assessing the impact of co-occurring mutations. We performed a retrospective cohort analysis of patients treated with HMA+VEN-based regimens at The University of Texas MD Anderson Cancer Center. A total of 119 patients (spliceosome mutated n = 39 [SRSF2, n = 24; SF3B1, n = 8; U2AF1, n = 7]; wild-type, n = 80) were included. Similar responses were observed between spliceosome and wild-type cohorts for composite complete response (CRc; 79% vs 75%, P = .65), and measurable residual disease-negative CRc (48% vs 60%, P = .34). Median overall survival for spliceosome vs wild-type patients was 35 vs 14 months (P = .58), and was not reached; 35 months and 8 months for patients with SRSF2, SF3B1, and U2AF1 mutations, respectively. IDH2 mutations were enriched in patients with SRSF2 mutations and associated with favorable outcomes (1- and 2-year overall survival [OS] of 100% and 88%). RAS mutations were enriched in patients with U2AF1 mutations and associated with inferior outcomes (median OS, 8 months). Comparable outcomes were observed between patients with vs without spliceosome mutations treated with HMA+VEN regimens, with specific co-mutation pairs demonstrating favorable outcomes.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: C.D.D. reports research support from AbbVie, Agios, Calithera, Cleave, BMS/Celgene, Daiichi-Sankyo, ImmuneOnc, and Loxo, and consultancy/advisory contributions for AbbVie, Agios, Celgene/BMS, Foghorn Therapeutics, Gilead, ImmuneOnc, Novartis, Takeda, and Notable Labs. N.J.S. reports research support from Takeda Oncology and Astellas Pharma Inc, consultancy support from Takeda Oncology and AstraZeneca, and honoraria from Amgen. N.D. reports research support from Daiichi-Sankyo, Bristol-Myers Squibb, Pfizer, Gilead, Sevier, Genentech, Astellas, Daiichi-Sankyo, AbbVie, Hanmi, Trovagene, FATE, Amgen, Novimmune, Glycomimetics, and ImmunoGen, and consultancy/advisory contributions to Daiichi-Sankyo, Bristol-Myers Squibb, Pfizer, Novartis, Celgene, AbbVie, Astellas, Genentech, Immunogen, Servier, Syndax, Trillium, Gilead, Amgen, and Agios. F.R. reports research support from AbbVie and advisory contributions and honoraria from Celgene and BMS. A.M. reports institutional research support from Celgene. T.K. reports research support from AbbVie, BMS, AstraZeneca, Cellenkos, Pfizer, Astellas, Ascentage, Pulmotec, Amgen, Genentech, Celgene, Incyte, Jazz, and Cyclacel, and honoraria from AbbVie, BMS, Pfizer, Novartis, Genentech, and Jazz. G.B. reports research contributions from AstraZeneca, GSK, BMS, AbbVie, Novartis, Incyte, Polaris, Xbiotech USA, Oncoceutics, PTC Therapeutics, Jannsen, Bioline Rx, Cyclacel, and consultancy contributions from PTC Therapeutics, Nkarta Therapeutics, Treadwell Therapeutics, BioTherix, Curio Science LLC, FTC Therapeutics, Argenx, and BioLine Rx. N.P. reports research contributions from Affymetrix, Stemline Therapeutics, AbbVie, Daiichi Sankyo, Plexxikon, Cellectis, Novartis, Samus Therapeutics; grant support from Affymetrix, SagerStrong; and honoraria from Stemline Therapeutics, LFB Biotechnologies, MustangBio, Incyte Corporation, Celgene, DAVA Oncology, Blueprint Medicines, Novartis, and Roche Diagnostics. K.S. reports research contributions from Novartis; consultancy contributions for Pfizer Japan, Novartis, Daiichi Sankyo; and honoraria from Otsuka. M.Y. reports research funding from Pfizer and Daiichi-Sankyo and honoraria from Pint Pharma. E.J. reports research contributions from Amgen, Adaptive Biotechnologies, AbbVie, Pfizer, Takeda, BMS, and Genentech, and advisory contributions for Amge, Adaptive Biotechnologies, AbbVie, Pfizer, Takeda, BMS, and Genentech. H.M.K. reports research contributions from AbbVie, Amgen, Jazz, Daiichi-Sankyo, BMS, Ascentage, Immunogen, Novartis, Pfizer, and Sanofi; advisory contributions for Actinium; and honoraria contributions from Aptitute Health, AbbVie, BioAscend, Δ Fly, Amgen, Daiichi-Sankyo, Adaptive Biotechnologies, Oxford Biomedical, Novartis, Actinium, and Pfizer. G.G.-M. reports research contributions from Celgene, Genentech, Novartis, Merck, Bristol-Myers Squibb, AbbVie, Helsinn Therapeutics, H3 Biomedicine, Amphivena Therapeutics, Onconova, Astex Pharmaceuticals; consultancy contributions from Celgene, Genentech, Bristol-Myers Squibb, Acceleron Pharmaceuticals, Helsinn Therapeutics, Astex Pharmaceuticals, and Jazz Pharmaceuticals; and honoraria contributions from Celgene, Acceleron Pharmaceuticals, AbbVie, Helsinn Therapeutics, and Astex Pharmaceuticals. M.Y.K. reports research contributions from Cellectis, Stemline Therapeutics, Ablynx, Forty-Seven, Genentech, Rafael Pharmaceutical, Agios, AstraZeneca, Calithera, Ascentage, AbbVie, Sanofi, Eli Lilly, F. Hoffmann-La Roche, and consultancy contributions to Stemline Therapeutics, Forty-Seven, Genentech, Kosji, AbbVie, Amgen, and F. Hoffmann-La Roche. The remaining authors declare no competing financial interests.
Figures


References
-
- Yoshida K, Sanada M, Shiraishi Y, et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. - PubMed
-
- Reinig E, Yang F, Traer E, et al. . Targeted next-generation sequencing in myelodysplastic syndrome and chronic myelomonocytic leukemia aids diagnosis in challenging cases and identifies frequent spliceosome mutations in transformed acute myeloid leukemia. Am J Clin Pathol. 2016;145(4):497-506. - PubMed
-
- Thol F, Kade S, Schlarmann C, et al. . Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578-3584. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

