A nucleotide-sensing endonuclease from the Gabija bacterial defense system
- PMID: 33885789
- PMCID: PMC8136825
- DOI: 10.1093/nar/gkab277
A nucleotide-sensing endonuclease from the Gabija bacterial defense system
Abstract
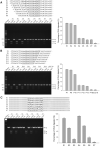
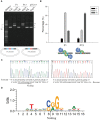
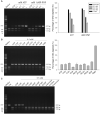
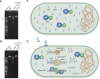
The arms race between bacteria and phages has led to the development of exquisite bacterial defense systems including a number of uncharacterized systems distinct from the well-known restriction-modification and CRISPR/Cas systems. Here, we report functional analyses of the GajA protein from the newly predicted Gabija system. The GajA protein is revealed as a sequence-specific DNA nicking endonuclease unique in that its activity is strictly regulated by nucleotide concentration. NTP and dNTP at physiological concentrations can fully inhibit the robust DNA cleavage activity of GajA. Interestingly, the nucleotide inhibition is mediated by an ATPase-like domain, which usually hydrolyzes ATP to stimulate the DNA cleavage when associated with other nucleases. These features suggest a mechanism of the Gabija defense in which an endonuclease activity is suppressed under normal conditions, while it is activated by the depletion of NTP and dNTP upon the replication and transcription of invading phages. This work highlights a concise strategy to utilize a DNA nicking endonuclease for phage resistance via nucleotide regulation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Dy R.L., Richter C., Salmond G.P.C., Fineran P.C.. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 2014; 1:307–331. - PubMed
-
- Labrie S.J., Samson J.E., Moineau S.. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010; 8:317–327. - PubMed
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau R., S. D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. - PubMed
-
- Tock M.R., Dryden D.T.F.. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005; 8:466–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

