iBLAST: Incremental BLAST of new sequences via automated e-value correction
- PMID: 33886589
- PMCID: PMC8062096
- DOI: 10.1371/journal.pone.0249410
iBLAST: Incremental BLAST of new sequences via automated e-value correction
Abstract
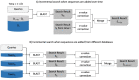
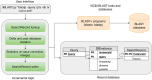
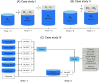
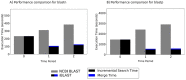
Search results from local alignment search tools use statistical scores that are sensitive to the size of the database to report the quality of the result. For example, NCBI BLAST reports the best matches using similarity scores and expect values (i.e., e-values) calculated against the database size. Given the astronomical growth in genomics data throughout a genomic research investigation, sequence databases grow as new sequences are continuously being added to these databases. As a consequence, the results (e.g., best hits) and associated statistics (e.g., e-values) for a specific set of queries may change over the course of a genomic investigation. Thus, to update the results of a previously conducted BLAST search to find the best matches on an updated database, scientists must currently rerun the BLAST search against the entire updated database, which translates into irrecoverable and, in turn, wasted execution time, money, and computational resources. To address this issue, we devise a novel and efficient method to redeem past BLAST searches by introducing iBLAST. iBLAST leverages previous BLAST search results to conduct the same query search but only on the incremental (i.e., newly added) part of the database, recomputes the associated critical statistics such as e-values, and combines these results to produce updated search results. Our experimental results and fidelity analyses show that iBLAST delivers search results that are identical to NCBI BLAST at a substantially reduced computational cost, i.e., iBLAST performs (1 + δ)/δ times faster than NCBI BLAST, where δ represents the fraction of database growth. We then present three different use cases to demonstrate that iBLAST can enable efficient biological discovery at a much faster speed with a substantially reduced computational cost.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature methods. 2014;12(1):59. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

