Some conditions apply: Systems for studying Plasmodium falciparum protein function
- PMID: 33886685
- PMCID: PMC8062007
- DOI: 10.1371/journal.ppat.1009442
Some conditions apply: Systems for studying Plasmodium falciparum protein function
Abstract
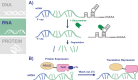
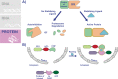
Malaria, caused by infection with Plasmodium parasites, remains a significant global health concern. For decades, genetic intractability and limited tools hindered our ability to study essential proteins and pathways in Plasmodium falciparum, the parasite associated with the most severe malaria cases. However, recent years have seen major leaps forward in the ability to genetically manipulate P. falciparum parasites and conditionally control protein expression/function. The conditional knockdown systems used in P. falciparum target all 3 components of the central dogma, allowing researchers to conditionally control gene expression, translation, and protein function. Here, we review some of the common knockdown systems that have been adapted or developed for use in P. falciparum. Much of the work done using conditional knockdown approaches has been performed in asexual, blood-stage parasites, but we also highlight their uses in other parts of the life cycle and discuss new ways of applying these systems outside of the intraerythrocytic stages. With the use of these tools, the field's understanding of parasite biology is ever increasing, and promising new pathways for antimalarial drug development are being discovered.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- World Health Organization. World malaria report 2020: 20 years of global progress and challenges. 2020;299.
-
- Meissner M, Krejany E, Gilson PR, de Koning-Ward TF, Soldati D, Crabb BS. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci U S A. 2005. February 22;102(8):2980–5. 10.1073/pnas.0500112102 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

