Structural Insights into the Mechanisms and Pharmacology of K2P Potassium Channels
- PMID: 33887333
- PMCID: PMC8436263
- DOI: 10.1016/j.jmb.2021.166995
Structural Insights into the Mechanisms and Pharmacology of K2P Potassium Channels
Abstract
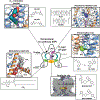
Leak currents, defined as voltage and time independent flows of ions across cell membranes, are central to cellular electrical excitability control. The K2P (KCNK) potassium channel class comprises an ion channel family that produces potassium leak currents that oppose excitation and stabilize the resting membrane potential in cells in the brain, cardiovascular system, immune system, and sensory organs. Due to their widespread tissue distribution, K2Ps contribute to many physiological and pathophysiological processes including anesthesia, pain, arrythmias, ischemia, hypertension, migraine, intraocular pressure regulation, and lung injury responses. Structural studies of six homomeric K2Ps have established the basic architecture of this channel family, revealed key moving parts involved in K2P function, uncovered the importance of asymmetric pinching and dilation motions in the K2P selectivity filter (SF) C-type gate, and defined two K2P structural classes based on the absence or presence of an intracellular gate. Further, a series of structures characterizing K2P:modulator interactions have revealed a striking polysite pharmacology housed within a relatively modestly sized (~70 kDa) channel. Binding sites for small molecules or lipids that control channel function are found at every layer of the channel structure, starting from its extracellular side through the portion that interacts with the membrane bilayer inner leaflet. This framework provides the basis for understanding how gating cues sensed by different channel parts control function and how small molecules and lipids modulate K2P activity. Such knowledge should catalyze development of new K2P modulators to probe function and treat a wide range of disorders.
Keywords: K(2P) channel; M2 and M4 helix transmembrane motions; polysite pharmacology; selectivity filter C-type gate.
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

