SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity
- PMID: 33888467
- PMCID: PMC8163077
- DOI: 10.1126/sciadv.abg7607
SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity
Abstract
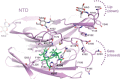
The coronaviral spike is the dominant viral antigen and the target of neutralizing antibodies. We show that SARS-CoV-2 spike binds biliverdin and bilirubin, the tetrapyrrole products of heme metabolism, with nanomolar affinity. Using cryo-electron microscopy and x-ray crystallography, we mapped the tetrapyrrole interaction pocket to a deep cleft on the spike N-terminal domain (NTD). At physiological concentrations, biliverdin significantly dampened the reactivity of SARS-CoV-2 spike with immune sera and inhibited a subset of neutralizing antibodies. Access to the tetrapyrrole-sensitive epitope is gated by a flexible loop on the distal face of the NTD. Accompanied by profound conformational changes in the NTD, antibody binding requires relocation of the gating loop, which folds into the cleft vacated by the metabolite. Our results indicate that SARS-CoV-2 spike NTD harbors a dominant epitope, access to which can be controlled by an allosteric mechanism that is regulated through recruitment of a metabolite.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures




Update of
-
SARS-CoV-2 recruits a haem metabolite to evade antibody immunity.medRxiv [Preprint]. 2021 Jan 26:2021.01.21.21249203. doi: 10.1101/2021.01.21.21249203. medRxiv. 2021. Update in: Sci Adv. 2021 May 28;7(22):eabg7607. doi: 10.1126/sciadv.abg7607. PMID: 33532784 Free PMC article. Updated. Preprint.
References
-
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C. J., Cerikan B., Lu J. M., Peukes J., Xiong X., Krausslich H. G., Scheres S. H. W., Bartenschlager R., Briggs J. A. G., Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). - PMC - PubMed
-
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

