Comprehensive characterization of tissue-specific chromatin accessibility in L2 Caenorhabditis elegans nematodes
- PMID: 33888511
- PMCID: PMC8494234
- DOI: 10.1101/gr.271791.120
Comprehensive characterization of tissue-specific chromatin accessibility in L2 Caenorhabditis elegans nematodes
Abstract
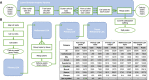
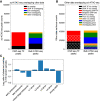
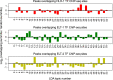
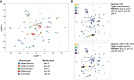
Recently developed single-cell technologies allow researchers to characterize cell states at ever greater resolution and scale. Caenorhabditis elegans is a particularly tractable system for studying development, and recent single-cell RNA-seq studies characterized the gene expression patterns for nearly every cell type in the embryo and at the second larval stage (L2). Gene expression patterns give insight about gene function and into the biochemical state of different cell types; recent advances in other single-cell genomics technologies can now also characterize the regulatory context of the genome that gives rise to these gene expression levels at a single-cell resolution. To explore the regulatory DNA of individual cell types in C. elegans, we collected single-cell chromatin accessibility data using the sci-ATAC-seq assay in L2 larvae to match the available single-cell RNA-seq data set. By using a novel implementation of the latent Dirichlet allocation algorithm, we identify 37 clusters of cells that correspond to different cell types in the worm, providing new maps of putative cell type-specific gene regulatory sites, with promise for better understanding of cellular differentiation and gene regulation.
© 2021 Durham et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





Similar articles
-
Ribosome biogenesis disruption mediated chromatin structure changes revealed by SRAtac, a customizable end to end analysis pipeline for ATAC-seq.BMC Genomics. 2023 Sep 1;24(1):512. doi: 10.1186/s12864-023-09576-y. BMC Genomics. 2023. PMID: 37658321 Free PMC article.
-
Genome-wide discovery of active regulatory elements and transcription factor footprints in Caenorhabditis elegans using DNase-seq.Genome Res. 2017 Dec;27(12):2108-2119. doi: 10.1101/gr.223735.117. Epub 2017 Oct 26. Genome Res. 2017. PMID: 29074739 Free PMC article.
-
Chromatin Immunoprecipitation and Sequencing (ChIP-seq) Optimized for Application in Caenorhabditis elegans.Curr Protoc. 2021 Jul;1(7):e187. doi: 10.1002/cpz1.187. Curr Protoc. 2021. PMID: 34242493
-
Modern techniques for the analysis of chromatin and nuclear organization in C. elegans.WormBook. 2014 Apr 2:1-35. doi: 10.1895/wormbook.1.169.1. WormBook. 2014. PMID: 24706462 Free PMC article. Review.
-
Systems biology of embryonic development: Prospects for a complete understanding of the Caenorhabditis elegans embryo.Wiley Interdiscip Rev Dev Biol. 2018 May;7(3):e314. doi: 10.1002/wdev.314. Epub 2018 Jan 25. Wiley Interdiscip Rev Dev Biol. 2018. PMID: 29369536 Review.
Cited by
-
Matrix prior for data transfer between single cell data types in latent Dirichlet allocation.PLoS Comput Biol. 2023 May 5;19(5):e1011049. doi: 10.1371/journal.pcbi.1011049. eCollection 2023 May. PLoS Comput Biol. 2023. PMID: 37146053 Free PMC article.
-
Modeling transcriptional regulation of model species with deep learning.Genome Res. 2021 Jun;31(6):1097-1105. doi: 10.1101/gr.266171.120. Epub 2021 Apr 22. Genome Res. 2021. PMID: 33888512 Free PMC article.
-
Hand Dissection of Caenorhabditis elegans Intestines.J Vis Exp. 2022 Sep 13;(187):10.3791/64120. doi: 10.3791/64120. J Vis Exp. 2022. PMID: 36190277 Free PMC article.
-
Detecting gene expression in Caenorhabditis elegans.Genetics. 2025 Jan 8;229(1):1-108. doi: 10.1093/genetics/iyae167. Genetics. 2025. PMID: 39693264 Free PMC article. Review.
-
Sensory plasticity caused by up-down regulation encodes the information of short-term learning and memory.iScience. 2025 Mar 13;28(4):112215. doi: 10.1016/j.isci.2025.112215. eCollection 2025 Apr 18. iScience. 2025. PMID: 40224011 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
