Modeling transcriptional regulation of model species with deep learning
- PMID: 33888512
- PMCID: PMC8168591
- DOI: 10.1101/gr.266171.120
Modeling transcriptional regulation of model species with deep learning
Abstract
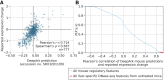
To enable large-scale analyses of transcription regulation in model species, we developed DeepArk, a set of deep learning models of the cis-regulatory activities for four widely studied species: Caenorhabditis elegans, Danio rerio, Drosophila melanogaster, and Mus musculus DeepArk accurately predicts the presence of thousands of different context-specific regulatory features, including chromatin states, histone marks, and transcription factors. In vivo studies show that DeepArk can predict the regulatory impact of any genomic variant (including rare or not previously observed) and enables the regulatory annotation of understudied model species.
© 2021 Cofer et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
