Genome-wide analysis of 944 133 individuals provides insights into the etiology of haemorrhoidal disease
- PMID: 33888516
- PMCID: PMC8292596
- DOI: 10.1136/gutjnl-2020-323868
Genome-wide analysis of 944 133 individuals provides insights into the etiology of haemorrhoidal disease
Abstract
Objective: Haemorrhoidal disease (HEM) affects a large and silently suffering fraction of the population but its aetiology, including suspected genetic predisposition, is poorly understood. We report the first genome-wide association study (GWAS) meta-analysis to identify genetic risk factors for HEM to date.
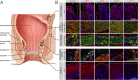
Design: We conducted a GWAS meta-analysis of 218 920 patients with HEM and 725 213 controls of European ancestry. Using GWAS summary statistics, we performed multiple genetic correlation analyses between HEM and other traits as well as calculated HEM polygenic risk scores (PRS) and evaluated their translational potential in independent datasets. Using functional annotation of GWAS results, we identified HEM candidate genes, which differential expression and coexpression in HEM tissues were evaluated employing RNA-seq analyses. The localisation of expressed proteins at selected loci was investigated by immunohistochemistry.
Results: We demonstrate modest heritability and genetic correlation of HEM with several other diseases from the GI, neuroaffective and cardiovascular domains. HEM PRS validated in 180 435 individuals from independent datasets allowed the identification of those at risk and correlated with younger age of onset and recurrent surgery. We identified 102 independent HEM risk loci harbouring genes whose expression is enriched in blood vessels and GI tissues, and in pathways associated with smooth muscles, epithelial and endothelial development and morphogenesis. Network transcriptomic analyses highlighted HEM gene coexpression modules that are relevant to the development and integrity of the musculoskeletal and epidermal systems, and the organisation of the extracellular matrix.
Conclusion: HEM has a genetic component that predisposes to smooth muscle, epithelial and connective tissue dysfunction.
Keywords: anal canal histopathology; anorectal disorders; genetics.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: Vladimir Vacic and Olga V. Sazonova are/were employed by and hold stock or stock options in 23andMe, Inc. All other authors have nothing to declare.
Figures



Comment in
-
Haemorrhoidal disease reduces the risk of diverticular disease and irritable bowel syndrome: a Mendelian randomisation study.Gut. 2024 Feb 23;73(3):553-554. doi: 10.1136/gutjnl-2022-329307. Gut. 2024. PMID: 36693712 No abstract available.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
