Conserved genetic signatures parcellate cardinal spinal neuron classes into local and projection subsets
- PMID: 33888637
- PMCID: PMC8612134
- DOI: 10.1126/science.abe0690
Conserved genetic signatures parcellate cardinal spinal neuron classes into local and projection subsets
Abstract
Motor and sensory functions of the spinal cord are mediated by populations of cardinal neurons arising from separate progenitor lineages. However, each cardinal class is composed of multiple neuronal types with distinct molecular, anatomical, and physiological features, and there is not a unifying logic that systematically accounts for this diversity. We reasoned that the expansion of new neuronal types occurred in a stepwise manner analogous to animal speciation, and we explored this by defining transcriptomic relationships using a top-down approach. We uncovered orderly genetic tiers that sequentially divide groups of neurons by their motor-sensory, local-long range, and excitatory-inhibitory features. The genetic signatures defining neuronal projections were tied to neuronal birth date and conserved across cardinal classes. Thus, the intersection of cardinal class with projection markers provides a unifying taxonomic solution for systematically identifying distinct functional subsets.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures



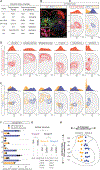

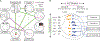
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

