Disruption of the HER3-PI3K-mTOR oncogenic signaling axis and PD-1 blockade as a multimodal precision immunotherapy in head and neck cancer
- PMID: 33888713
- PMCID: PMC8062674
- DOI: 10.1038/s41467-021-22619-w
Disruption of the HER3-PI3K-mTOR oncogenic signaling axis and PD-1 blockade as a multimodal precision immunotherapy in head and neck cancer
Abstract
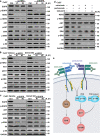
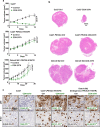
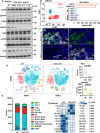
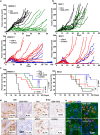
Immune checkpoint blockade (ICB) therapy has revolutionized head and neck squamous cell carcinoma (HNSCC) treatment, but <20% of patients achieve durable responses. Persistent activation of the PI3K/AKT/mTOR signaling circuitry represents a key oncogenic driver in HNSCC; however, the potential immunosuppressive effects of PI3K/AKT/mTOR inhibitors may limit the benefit of their combination with ICB. Here we employ an unbiased kinome-wide siRNA screen to reveal that HER3, is essential for the proliferation of most HNSCC cells that do not harbor PIK3CA mutations. Indeed, we find that persistent tyrosine phosphorylation of HER3 and PI3K recruitment underlies aberrant PI3K/AKT/mTOR signaling in PIK3CA wild type HNSCCs. Remarkably, antibody-mediated HER3 blockade exerts a potent anti-tumor effect by suppressing HER3-PI3K-AKT-mTOR oncogenic signaling and concomitantly reversing the immune suppressive tumor microenvironment. Ultimately, we show that HER3 inhibition and PD-1 blockade may provide a multimodal precision immunotherapeutic approach for PIK3CA wild type HNSCC, aimed at achieving durable cancer remission.
Conflict of interest statement
J.S.G. has received other commercial research support from Kura Oncology and Mavupharma, and is a consultant/advisory board member for Oncoceutics Inc., Vividion Therapeutics, and Domain Therapeutics; The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE030497/DE/NIDCR NIH HHS/United States
- F32 DE029990/DE/NIDCR NIH HHS/United States
- R01 DE026644/DE/NIDCR NIH HHS/United States
- S10 OD021831/OD/NIH HHS/United States
- R01 DE023685/DE/NIDCR NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- T32 GM007752/GM/NIGMS NIH HHS/United States
- U01 DE028227/DE/NIDCR NIH HHS/United States
- R01 CA247551/CA/NCI NIH HHS/United States
- R01 DE026870/DE/NIDCR NIH HHS/United States
- U54 CA209891/CA/NCI NIH HHS/United States
- R01 DE028343/DE/NIDCR NIH HHS/United States
- R35 CA231998/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

