BRCA1 degradation in response to mitochondrial damage in breast cancer cells
- PMID: 33888730
- PMCID: PMC8062582
- DOI: 10.1038/s41598-021-87698-7
BRCA1 degradation in response to mitochondrial damage in breast cancer cells
Abstract
BRCA1 is a well-studied tumor suppressor involved in the homologous repair of DNA damage, whereas PINK1, a mitochondrial serine/threonine kinase, is known to be involved in mitochondrial quality control. Genetic mutations of PINK1 and Parkin cause autosomal recessive early-onset Parkinson's disease. We found that in breast cancer cells, the mitochondrial targeting reagents, which all induce mitochondrial depolarization along with PINK1 upregulation, induced proteasomal BRCA1 degradation. This BRCA1 degradation was dependent on PINK1, and BRCA1 downregulation upon mitochondrial damage caused DNA double-strand breaks. BRCA1 degradation was mediated through the direct interaction with the E3 ligase Parkin. Strikingly, BRCA1 and PINK1/Parkin expression were inversely correlated in cancerous mammary glands from breast cancer patients. BRCA1 knockdown repressed cancer cell growth, and high BRCA1 expression predicted poor relapse-free survival in breast cancer patients. These observations indicate a novel mechanism by which mitochondrial damage is transmitted to the nucleus, leading to BRCA1 degradation.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
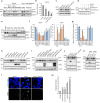

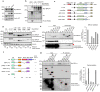

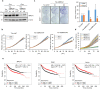
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

