MultiVERSE: a multiplex and multiplex-heterogeneous network embedding approach
- PMID: 33888761
- PMCID: PMC8062697
- DOI: 10.1038/s41598-021-87987-1
MultiVERSE: a multiplex and multiplex-heterogeneous network embedding approach
Abstract
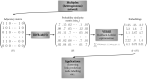
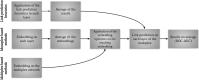
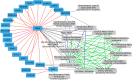
Network embedding approaches are gaining momentum to analyse a large variety of networks. Indeed, these approaches have demonstrated their effectiveness in tasks such as community detection, node classification, and link prediction. However, very few network embedding methods have been specifically designed to handle multiplex networks, i.e. networks composed of different layers sharing the same set of nodes but having different types of edges. Moreover, to our knowledge, existing approaches cannot embed multiple nodes from multiplex-heterogeneous networks, i.e. networks composed of several multiplex networks containing both different types of nodes and edges. In this study, we propose MultiVERSE, an extension of the VERSE framework using Random Walks with Restart on Multiplex (RWR-M) and Multiplex-Heterogeneous (RWR-MH) networks. MultiVERSE is a fast and scalable method to learn node embeddings from multiplex and multiplex-heterogeneous networks. We evaluate MultiVERSE on several biological and social networks and demonstrate its performance. MultiVERSE indeed outperforms most of the other methods in the tasks of link prediction and network reconstruction for multiplex network embedding, and is also efficient in link prediction for multiplex-heterogeneous network embedding. Finally, we apply MultiVERSE to study rare disease-gene associations using link prediction and clustering. MultiVERSE is freely available on github at https://github.com/Lpiol/MultiVERSE .
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Hamilton, W. L. & Ying, R., Leskovec, J. Methods and applications. IEEE Data Engineering Bulletin, Representation learning on graphs, (2017).
-
- Liao L, He X, Zhang H, Chua T-S. Attributed social network embedding. IEEE Trans. Knowl. Data Eng. 2018;30:2257–2270. doi: 10.1109/TKDE.2018.2819980. - DOI
-
- Ma, G., Lu, C.-T., He, L., Philip, S. Y. & Ragin, A. B. Multi-view graph embedding with hub detection for brain network analysis. In 2017 IEEE International Conference on Data Mining (ICDM) 967–972 (IEEE, 2017).
-
- Perozzi, B., Al-Rfou, R. & Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining 701–710 (ACM, 2014).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

