Phosphoproteome Study of Escherichia coli Devoid of Ser/Thr Kinase YeaG During the Metabolic Shift From Glucose to Malate
- PMID: 33889145
- PMCID: PMC8055822
- DOI: 10.3389/fmicb.2021.657562
Phosphoproteome Study of Escherichia coli Devoid of Ser/Thr Kinase YeaG During the Metabolic Shift From Glucose to Malate
Abstract
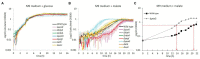
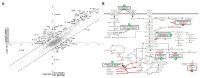
Understanding phosphorylation-mediated regulation of metabolic enzymes, pathways, and cell phenotypes under metabolic shifts represents a major challenge. The kinases associated with most phosphorylation sites and the link between phosphorylation and enzyme activity remain unknown. In this study, we performed stable isotope labeling by amino acids in cell culture (SILAC)-based proteome and phosphoproteome analysis of Escherichia coli ΔyeaG, a strain lacking a poorly characterized serine/threonine kinase YeaG, to decipher kinase-substrate interactions and the effects on metabolic phenotype during shifts from glucose to malate. The starting point of our analysis was the identification of physiological conditions under which ΔyeaG exhibits a clear phenotype. By metabolic profiling, we discovered that ΔyeaG strain has a significantly shorter lag phase than the wild type during metabolic shift from glucose to malate. Under those conditions, our SILAC analysis revealed several proteins that were differentially phosphorylated in the ΔyeaG strain. By focusing on metabolic enzymes potentially involved in central carbon metabolism, we narrowed down our search for putative YeaG substrates and identified isocitrate lyase AceA as the direct substrate of YeaG. YeaG was capable of phosphorylating AceA in vitro only in the presence of malate, suggesting that this phosphorylation event is indeed relevant for glucose to malate shift. There is currently not enough evidence to firmly establish the exact mechanism of this newly observed regulatory phenomenon. However, our study clearly exemplifies the usefulness of SILAC-based approaches in identifying proteins kinase substrates, when applied in physiological conditions relevant for the activity of the protein kinase in question.
Keywords: SILAC; kinase-substrate relationship; metabolic adaptation; phosphoproteome; protein kinase.
Copyright © 2021 Sultan, Jers, Ganief, Shi, Senissar, Køhler, Macek and Mijakovic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Cloning, expression, purification and characterization of the stress kinase YeaG from Escherichia coli.Protein Expr Purif. 2008 May;59(1):79-85. doi: 10.1016/j.pep.2008.01.005. Epub 2008 Jan 24. Protein Expr Purif. 2008. PMID: 18276156
-
Control of isocitrate dehydrogenase catalytic activity by protein phosphorylation in Escherichia coli.J Mol Microbiol Biotechnol. 2005;9(3-4):132-46. doi: 10.1159/000089642. J Mol Microbiol Biotechnol. 2005. PMID: 16415587 Review.
-
Global dynamics of the Escherichia coli proteome and phosphoproteome during growth in minimal medium.J Proteome Res. 2013 Jun 7;12(6):2611-21. doi: 10.1021/pr3011843. Epub 2013 May 2. J Proteome Res. 2013. PMID: 23590516
-
Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes.Arch Microbiol. 1997 Oct;168(4):262-9. doi: 10.1007/s002030050497. Arch Microbiol. 1997. PMID: 9297462
-
Regulation of acetate metabolism by protein phosphorylation in enteric bacteria.Annu Rev Microbiol. 1998;52:127-64. doi: 10.1146/annurev.micro.52.1.127. Annu Rev Microbiol. 1998. PMID: 9891796 Review.
Cited by
-
Extensive regulation of enzyme activity by phosphorylation in Escherichia coli.Nat Commun. 2021 Sep 24;12(1):5650. doi: 10.1038/s41467-021-25988-4. Nat Commun. 2021. PMID: 34561442 Free PMC article.
-
Identification of serine/threonine kinases that regulate metabolism and sporulation in Clostridium beijerinckii.Appl Microbiol Biotechnol. 2022 Nov;106(22):7563-7575. doi: 10.1007/s00253-022-12234-0. Epub 2022 Oct 26. Appl Microbiol Biotechnol. 2022. PMID: 36287220
-
Experimental measurement and computational prediction of bacterial Hanks-type Ser/Thr signaling system regulatory targets.Mol Microbiol. 2024 Aug;122(2):152-164. doi: 10.1111/mmi.15220. Epub 2024 Jan 3. Mol Microbiol. 2024. PMID: 38167835 Free PMC article. Review.
-
The Role of Propionate-Induced Rearrangement of Membrane Proteins in the Formation of the Virulent Phenotype of Crohn's Disease-Associated Adherent-Invasive Escherichia coli.Int J Mol Sci. 2024 Sep 20;25(18):10118. doi: 10.3390/ijms251810118. Int J Mol Sci. 2024. PMID: 39337603 Free PMC article.
-
Spatio-temporal organization of the E. coli chromosome from base to cellular length scales.EcoSal Plus. 2024 Dec 12;12(1):eesp00012022. doi: 10.1128/ecosalplus.esp-0001-2022. Epub 2024 Jun 12. EcoSal Plus. 2024. PMID: 38864557 Free PMC article. Review.
References
-
- Arora G., Sajid A., Gupta M., Bhaduri A., Kumar P., Basu-Modak S., et al. . (2010). Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS One 5:e10772. 10.1371/journal.pone.0010772, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

