Analytic validation and clinical utilization of the comprehensive genomic profiling test, GEM ExTra®
- PMID: 33889297
- PMCID: PMC8057276
- DOI: 10.18632/oncotarget.27945
Analytic validation and clinical utilization of the comprehensive genomic profiling test, GEM ExTra®
Abstract
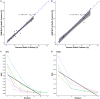
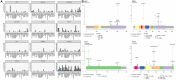
We developed and analytically validated a comprehensive genomic profiling (CGP) assay, GEM ExTra, for patients with advanced solid tumors that uses Next Generation Sequencing (NGS) to characterize whole exomes employing a paired tumor-normal subtraction methodology. The assay detects single nucleotide variants (SNV), indels, focal copy number alterations (CNA), TERT promoter region, as well as tumor mutation burden (TMB) and microsatellite instability (MSI) status. Additionally, the assay incorporates whole transcriptome sequencing of the tumor sample that allows for the detection of gene fusions and select special transcripts, including AR-V7, EGFR vIII, EGFRvIV, and MET exon 14 skipping events. The assay has a mean target coverage of 180X for the normal (germline) and 400X for tumor DNA including enhanced probe design to facilitate the sequencing of difficult regions. Proprietary bioinformatics, paired with comprehensive clinical curation results in reporting that defines clinically actionable, FDA-approved, and clinical trial drug options for the management of the patient's cancer. GEM ExTra demonstrated analytic specificity (PPV) of > 99.9% and analytic sensitivity of 98.8%. Application of GEM ExTra to 1,435 patient samples revealed clinically actionable alterations in 83.9% of reports, including 31 (2.5%) where therapeutic recommendations were based on RNA fusion findings only.
Keywords: RNA sequencing; comprehensive genomic profiling; pan-cancer; precision medicine; whole exome sequencing.
Copyright: © 2021 White et al.
Conflict of interest statement
CONFLICTS OF INTEREST All authors have a financial relationship as employees of Ashion Analytics.
Figures



References
-
- El-Deiry WS, Goldberg RM, Lenz HJ, Shields AF, Gibney GT, Tan AR, Brown J, Eisenberg B, Heath EI, Phuphanich S, Kim E, Brenner AJ, Marshall JL. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J Clin. 2019; 69:305–43. 10.3322/caac.21560. - DOI - PMC - PubMed
-
- Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, Subbiah V, Fu S, Karp D, Falchook GS, Tsimberidou AM, Piha-Paul S, Anderson R, et al.. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res. 2016; 76:3690–701. 10.1158/0008-5472.CAN-15-3043. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

