Differential Expression of Insulin-Like Growth Factor 1 and Wnt Family Member 4 Correlates With Functional Heterogeneity of Human Dermal Fibroblasts
- PMID: 33889572
- PMCID: PMC8056032
- DOI: 10.3389/fcell.2021.628039
Differential Expression of Insulin-Like Growth Factor 1 and Wnt Family Member 4 Correlates With Functional Heterogeneity of Human Dermal Fibroblasts
Abstract
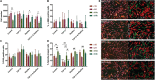
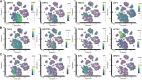
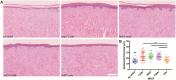
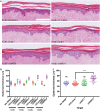
Although human dermis contains distinct fibroblast subpopulations, the functional heterogeneity of fibroblast lines from different donors is under-appreciated. We identified one commercially sourced fibroblast line (c64a) that failed to express α-smooth muscle actin (α-SMA), a marker linked to fibroblast contractility, even when treated with transforming growth factor-β1 (TGF-β1). Gene expression profiling identified insulin-like growth factor 1 (IGF1) as being expressed more highly, and Asporin (ASPN) and Wnt family member 4 (WNT4) expressed at lower levels, in c64a fibroblasts compared to three fibroblast lines that had been generated in-house, independent of TGF-β1 treatment. TGF-β1 increased expression of C-X-C motif chemokine ligand 1 (CXCL1) in c64a cells to a greater extent than in the other lines. The c64a gene expression profile did not correspond to any dermal fibroblast subpopulation identified by single-cell RNAseq of freshly isolated human skin cells. In skin reconstitution assays, c64a fibroblasts did not support epidermal stratification as effectively as other lines tested. In fibroblast lines generated in-house, shRNA-mediated knockdown of IGF1 increased α-SMA expression without affecting epidermal stratification. Conversely, WNT4 knockdown had no consistent effect on α-SMA expression, but increased the ability of fibroblasts to support epidermal stratification. Thus, by comparing the properties of different lines of cultured dermal fibroblasts, we have identified IGF1 and WNT4 as candidate mediators of two distinct dermal functions: myofibroblast formation and epidermal maintenance.
Keywords: IGF1; WNT4; cell culture; epidermis; fibroblasts; signalling; skin; α-SMA.
Copyright © 2021 Culley, Louis, Philippeos, Oulès, Tihy, Segal, Hyliands, Jenkins, Bhogal, Siow and Watt.
Conflict of interest statement
DH, GJ, and RB are employees of Unilever. FW is on secondment as Executive Chair, Medical Research Council. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Clark R. A., McCoy G. A., Folkvord J. M., McPherson J. M. (1997). TGF-beta 1 stimulates cultured human fibroblasts to proliferate and produce tissue-like fibroplasia: a fibronectin matrix-dependent event. J. Cell. Physiol. 170 69–80. 10.1002/(sici)1097-4652(199701)170:1<69::aid-jcp8>3.0.co;2-j - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

