Acidic pH Decreases the Endonuclease Activity of Initiator RepB and Increases the Stability of the Covalent RepB-DNA Intermediate while Has Only a Limited Effect on the Replication of Plasmid pMV158 in Lactococcus lactis
- PMID: 33889596
- PMCID: PMC8056398
- DOI: 10.3389/fmolb.2021.634461
Acidic pH Decreases the Endonuclease Activity of Initiator RepB and Increases the Stability of the Covalent RepB-DNA Intermediate while Has Only a Limited Effect on the Replication of Plasmid pMV158 in Lactococcus lactis
Abstract
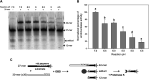
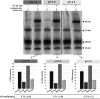
Plasmid vectors constitute a valuable tool for homologous and heterologous gene expression, for characterization of promoter and regulatory regions, and for genetic manipulation and labeling of bacteria. During the last years, a series of vectors based on promiscuous replicons of the pMV158 family have been developed for their employment in a variety of Gram-positive bacteria and proved to be useful for all above applications in lactic acid bacteria. A proper use of the plasmid vectors requires detailed knowledge of their main replicative features under the changing growth conditions of the studied bacteria, such as the acidification of the culture medium by lactic acid production. Initiation of pMV158 rolling-circle replication is catalyzed by the plasmid-encoded RepB protein, which performs a sequence-specific cleavage on one of the parental DNA strands and, as demonstrated in this work, establishes a covalent bond with the 5'-P end generated in the DNA. This covalent adduct must last until the leading-strand termination stage, where a new cleavage on the regenerated nick site and a subsequent strand-transfer reaction result in rejoining of the ends of the cleaved parental strand, whereas hydrolysis of the newly-generated adduct would release the protein from a nicked double-stranded DNA plasmid form. We have analyzed here the effect of pH on the different in vitro reactions catalyzed by RepB and on the in vivo replication ability of plasmid pMV158. We show that acidic pH greatly impairs the catalytic activity of the protein and reduces hydrolysis of the covalent RepB-DNA adduct, as expected for the nucleophilic nature of these reactions. Conversely, the ability of pMV158 to replicate in vivo, as monitored by the copy number and segregational stability of the plasmid in Lactococcus lactis, remains almost intact at extracellular pHs ranging from 7.0 to 5.0, and a significant reduction (by ∼50%) in the plasmid copy number per chromosome equivalent is only observed at pH 4.5. Moreover, the RepB to pMV158 molar ratio is increased at pH 4.5, suggesting the existence of compensatory mechanisms that operate in vivo to allow pMV158 replication at pH values that severely disturb the catalytic activity of the initiator protein.
Keywords: covalent RepB-DNA adduct; endonuclease activity; nucleophilic attack; promiscuous plasmid pMV158; rolling circle replication (RCR); strand-transfer activity.
Copyright © 2021 Valdelvira, Bordanaba-Ruiseco, Martín-Huestamendía, Ruiz-Masó and del Solar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Bordanaba-Ruiseco L. (2017). Caracterización de las actividades de RepB y su implicación en el proceso replicativo del plásmido promiscuo pMV158. Doctoral Thesis. Complutense University of Madrid. Available at: https://www.educacion.gob.es/teseo/mostrarRef.do?ref=433542
LinkOut - more resources
Full Text Sources
Other Literature Sources

