Characterization of the complete chloroplast genome of China Viburnum burejaeticum Regel et Herd and intra-species diversity
- PMID: 33889747
- PMCID: PMC8032331
- DOI: 10.1080/23802359.2021.1907810
Characterization of the complete chloroplast genome of China Viburnum burejaeticum Regel et Herd and intra-species diversity
Abstract
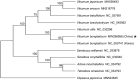
Viburnum burejaeticum Regel et Herd is widely cultivated in botanical gardens. However, as a member of Adoxaceae, few studies have been carried out on its phylogenetic relationship with other family members. Here we report the first complete chloroplast (cp) genome sequence of V. burejaeticum collected from China. The circular cp genome is 158,381 bp in size, including a large single-copy (LSC) region of 87,067 bp and a small single-copy (SSC) region of 18,212 bp, which were separated by two inverted repeat (IR) regions (26,551 bp each). A total of 126 genes were annotated, including 8 ribosomal RNAs (rRNAs), 36 transfer RNAs (tRNAs), and 82 protein-coding genes (PCGs). The sequence comparison of two V. burejaeticum collected from Korea and China revealed 101 single nucleotide polymorphisms (SNPs) and 16 insertions/deletions (InDels). In addition, maximum-likelihood (ML) phylogenetic analysis indicated V. burejaeticum species collected in Korea and China are clustered together. This study provides useful information for future genetic study of V. burejaeticum.
Keywords: Adoxaceae; Viburnum burejaeticum; complete chloroplast genome; phylogenetic.
© 2021 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
The complete chloroplast genome of Viburnum sargentii Koehne (Adoxaceae).Mitochondrial DNA B Resour. 2021 Apr 8;6(4):1328-1329. doi: 10.1080/23802359.2021.1910087. Mitochondrial DNA B Resour. 2021. PMID: 33889741 Free PMC article.
-
The complete chloroplast genome sequence of Viburnum Japonicum (Adoxaceae), an evergreen broad-leaved shrub.Mitochondrial DNA B Resour. 2018 Apr 12;3(1):458-459. doi: 10.1080/23802359.2018.1462121. Mitochondrial DNA B Resour. 2018. PMID: 33490513 Free PMC article.
-
The second complete chloroplast genome sequence of the Viburnum erosum (Adoxaceae) showed a low level of intra-species variations.Mitochondrial DNA B Resour. 2019 Dec 13;5(1):271-272. doi: 10.1080/23802359.2019.1698360. Mitochondrial DNA B Resour. 2019. PMID: 33366517 Free PMC article.
-
Characterization of the complete chloroplast genome of Viburnum schensianum (Adoxaceae).Mitochondrial DNA B Resour. 2020 Feb 28;5(2):1196-1197. doi: 10.1080/23802359.2020.1731357. eCollection 2020. Mitochondrial DNA B Resour. 2020. PMID: 33366910 Free PMC article.
-
The complete chloroplast genome of Lonicera hypoglauca Miq (Caprifoliaceae: Dipsacales) from Guangxi, China.Mitochondrial DNA B Resour. 2021 Feb 9;6(2):450-451. doi: 10.1080/23802359.2020.1870903. Mitochondrial DNA B Resour. 2021. PMID: 33659704 Free PMC article.
Cited by
-
Assembly and comparative analysis of the complete mitochondrial genome of Viburnum chinshanense.BMC Plant Biol. 2023 Oct 11;23(1):487. doi: 10.1186/s12870-023-04493-4. BMC Plant Biol. 2023. PMID: 37821817 Free PMC article.
References
-
- Clement WL, Arakaki M, Sweeney PW, Edwards EJ, Donoghue MJ.. 2014. A chloroplast tree for Viburnum (Adoxaceae) and its implications for phylogenetic classification and character evolution. Am J Bot. 101(6):1029–1049. - PubMed
-
- Nguyen VB, Linh Giang VN, Waminal NE, Park HS, Kim NH, Jang W, Lee J, Yang TJ.. 2020. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J Ginseng Res. 44(1):135–144. - PMC - PubMed
-
- Nguyen VB, Park HS, Lee SC, Lee J, Park JY, Yang TJ.. 2017. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J Agric Food Chem. 65(30):6298–6306. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
