Pharmacophore model, docking, QSAR, and molecular dynamics simulation studies of substituted cyclic imides and herbal medicines as COX-2 inhibitors
- PMID: 33889764
- PMCID: PMC8047494
- DOI: 10.1016/j.heliyon.2021.e06605
Pharmacophore model, docking, QSAR, and molecular dynamics simulation studies of substituted cyclic imides and herbal medicines as COX-2 inhibitors
Abstract
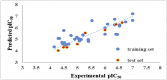
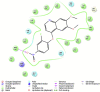
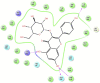
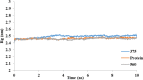
Cyclooxygenase-2 (COX-2) enzyme inhibitors have not eliminated the necessity for developed drugs not only in the nonsteroidal anti-inflammatory drug (NSAIDs) area, but also in other therapeutic applications including prevention of cancer and Alzheimer's disease. A series of novel substituted cyclic imides have been reported as selective COX-2 inhibitors. To understand the structural features responsible for their activity, a 3D validated pharmacophore and quantitative structure-activity relationship (QSAR) model have been developed. The values of enrichment factor (EF), goodness of hit score (GH), area under the ROC curve (AUC), sensitivity, and specificity refer to the good ability of the pharmacophore model to identify active compounds. Multiple linear regression (MLR) produced statistically significant QSAR model with (R2 training = 0.763, R2 test = 0.96) and predictability (Q2 training = 0.66, Q2 test = 0.84). Then, using the pharmacophore and QSAR models, eight authenticated botanicals in two herbal medicines and the ZINC compounds database, were virtually screened for ligands to COX-2. The retrieved hits which also obey lipinski's rule of five (RO5) were docked in the COX-2 3D structure to investigate their binding mode and affinity. Finally, based on the docking results, nine molecules were prioritized as promising hits that could be used as leads to discover novel COX-2 inhibitors. COX-2 inhibition of most of these hits has not been reported previously. Ten-nanosecond molecular dynamics simulation (10-ns MD) was performed on the initial structure COX-2 complex with ZINC000113253375 and ZINC000043170560 resulted from the docking. Our utilization of the 3D pharmacophore model, QSAR, molecular docking, and molecular dynamics simulation trials can be a potent strategy to successfully predict activity, efficiently design drugs, and screen large numbers of new compounds as active drug candidates.
Keywords: Cyclooxygenase-2; Docking; Molecular dynamics simulation; Pharmacophore; QSAR; Virtual screening.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
In Silico Drug-designing Studies on Sulforaphane Analogues: Pharmacophore Mapping, Molecular Docking and QSAR Modeling.Curr Drug Discov Technol. 2021;18(1):139-157. doi: 10.2174/1570163816666191112122047. Curr Drug Discov Technol. 2021. PMID: 31721705
-
Pharmacophore generation, atom-based 3D-QSAR, molecular docking and molecular dynamics simulation studies on benzamide analogues as FtsZ inhibitors.J Biomol Struct Dyn. 2018 Sep;36(12):3218-3230. doi: 10.1080/07391102.2017.1384401. Epub 2017 Oct 11. J Biomol Struct Dyn. 2018. PMID: 28938860
-
Structural Investigation of Vinca Domain Tubulin Binders by Pharmacophore, Atom based QSAR, Docking and Molecular Dynamics Simulations.Comb Chem High Throughput Screen. 2017;20(8):682-695. doi: 10.2174/1386207320666170509151253. Comb Chem High Throughput Screen. 2017. PMID: 28486912
-
Identification of Potent Small-Molecule PCSK9 Inhibitors Based on Quantitative Structure-Activity Relationship, Pharmacophore Modeling, and Molecular Docking Procedure.Curr Probl Cardiol. 2023 Jun;48(6):101660. doi: 10.1016/j.cpcardiol.2023.101660. Epub 2023 Feb 24. Curr Probl Cardiol. 2023. PMID: 36841313 Review.
-
Detection of Natural Inhibitors against Human Liver Cancer Cell Lines through QSAR, Molecular Docking and ADMET Studies.Curr Top Med Chem. 2021;21(8):686-695. doi: 10.2174/1568026620666201204155830. Curr Top Med Chem. 2021. PMID: 33280598 Review.
Cited by
-
Ligand based pharmacophore modelling and integrated computational approaches in the quest for small molecule inhibitors against hCA IX.RSC Adv. 2024 Jan 22;14(5):3346-3358. doi: 10.1039/d3ra08618f. eCollection 2024 Jan 17. RSC Adv. 2024. PMID: 38259989 Free PMC article.
-
LIGHTHOUSE illuminates therapeutics for a variety of diseases including COVID-19.iScience. 2022 Oct 10;25(11):105314. doi: 10.1016/j.isci.2022.105314. eCollection 2022 Nov 18. iScience. 2022. PMID: 36246574 Free PMC article.
-
N-alkylation of amines for the synthesis of potential antiviral agents: A structural modification approach.Heliyon. 2024 Sep 27;10(19):e38587. doi: 10.1016/j.heliyon.2024.e38587. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397970 Free PMC article.
-
Unveiling Novel Arginase Inhibitors for Cutaneous Leishmaniasis Using Drug Repurposing and Virtual Screening Approaches.J Cell Biochem. 2025 Aug;126(8):e70060. doi: 10.1002/jcb.70060. J Cell Biochem. 2025. PMID: 40847551 Free PMC article.
-
Anti-Inflammatory Effect of Izalpinin Derived from Chromolaena leivensis: λ-Carrageenan-Induced Paw Edema and In Silico Model.Molecules. 2023 Apr 26;28(9):3722. doi: 10.3390/molecules28093722. Molecules. 2023. PMID: 37175132 Free PMC article.
References
-
- Abouzid K., Frohberg P., Lehmann J., Decker M. 6-Aryl-4-oxohexanoic acids: synthesis, effects on eicosanoid biosynthesis, and anti-inflammatory in vivo-activities. Med. Chem. 2007;3:433–438. - PubMed
-
- Ulbrich H., Fiebich B., Dannhardt G. Cyclooxygenase-1/2 (COX-1/COX-2) and 5-lipoxygenase (5-LOX) inhibitors of the 6,7-diaryl-2,3-1H-dihydropyrrolizine type. ChemInform. 2003;34(20):953–959. - PubMed
-
- Dannhardt G., Laufer S. Structural approaches to explain the selectivity of COX-2 inhibitors: is there a common pharmacophore? Curr. Med. Chem. 2000;7(11):1101–1112. - PubMed
-
- Kim H.-J., Chae C.H., Yi K.Y., Park K.-L., Yoo S. Computational studies of COX-2 inhibitors: 3D-QSAR and docking. Bioorg. Med. Chem. 2004;12(7):1629–1641. - PubMed
-
- Almansa C., de Arriba A.F., Cavalcanti F.L., Gómez L.A., Miralles A., Merlos M., Forn J. Synthesis and SAR of a new series of COX-2-selective inhibitors: pyrazolo [1,5-a] pyrimidines. J. Med. Chem. 2001;44(3):350–361. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

