SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation
- PMID: 33890572
- PMCID: PMC8104966
- DOI: 10.7554/eLife.65962
SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation
Abstract
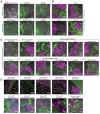
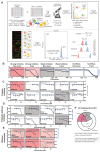
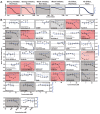
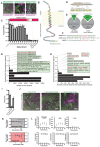
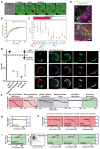
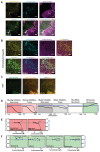
Many enveloped viruses induce multinucleated cells (syncytia), reflective of membrane fusion events caused by the same machinery that underlies viral entry. These syncytia are thought to facilitate replication and evasion of the host immune response. Here, we report that co-culture of human cells expressing the receptor ACE2 with cells expressing SARS-CoV-2 spike, results in synapse-like intercellular contacts that initiate cell-cell fusion, producing syncytia resembling those we identify in lungs of COVID-19 patients. To assess the mechanism of spike/ACE2-driven membrane fusion, we developed a microscopy-based, cell-cell fusion assay to screen ~6000 drugs and >30 spike variants. Together with quantitative cell biology approaches, the screen reveals an essential role for biophysical aspects of the membrane, particularly cholesterol-rich regions, in spike-mediated fusion, which extends to replication-competent SARS-CoV-2 isolates. Our findings potentially provide a molecular basis for positive outcomes reported in COVID-19 patients taking statins and suggest new strategies for therapeutics targeting the membrane of SARS-CoV-2 and other fusogenic viruses.
Keywords: ACE2; COVID-19; SARS-CoV-2; cell biology; coronavirus; human; spike; syncytia.
© 2021, Sanders et al.
Conflict of interest statement
DS, CJ, PA, DB, AD, HK, DK, IC, SS, TT, AT, MS, AP, IL, FD, RP, BL No competing interests declared, AH ASH is a consultant for Dewpoint Therapeutics. CB CPB is a scientific founder and consultant for Nereid Therapeutics.
Figures







References
-
- Abrams ME, Johnson KA, Perelman SS, Zhang LS, Endapally S, Mar KB, Thompson BM, McDonald JG, Schoggins JW, Radhakrishnan A, Alto NM. Oxysterols provide innate immunity to bacterial infection by mobilizing cell surface accessible cholesterol. Nature Microbiology. 2020;5:929–942. doi: 10.1038/s41564-020-0701-5. - DOI - PMC - PubMed
-
- Bauer L, Ferla S, Head SA, Bhat S, Pasunooti KK, Shi WQ, Albulescu L, Liu JO, Brancale A, van Kuppeveld FJM, Strating J. Structure-activity relationship study of itraconazole, a broad-range inhibitor of Picornavirus replication that targets oxysterol-binding protein (OSBP) Antiviral Research. 2018;156:55–63. doi: 10.1016/j.antiviral.2018.05.010. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

