A single point mutation converts a glutaryl-7-aminocephalosporanic acid acylase into an N-acyl-homoserine lactone acylase
- PMID: 33891232
- PMCID: PMC8197700
- DOI: 10.1007/s10529-021-03135-9
A single point mutation converts a glutaryl-7-aminocephalosporanic acid acylase into an N-acyl-homoserine lactone acylase
Abstract
Objective: To change the specificity of a glutaryl-7-aminocephalosporanic acid acylase (GCA) towards N-acyl homoserine lactones (AHLs; quorum sensing signalling molecules) by site-directed mutagenesis.
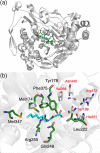
Results: Seven residues were identified by analysis of existing crystal structures as potential determinants of substrate specificity. Site-saturation mutagenesis libraries were created for each of the seven selected positions. High-throughput activity screening of each library identified two variants-Arg255Ala, Arg255Gly-with new activities towards N-acyl homoserine lactone substrates. Structural modelling of the Arg255Gly mutation suggests that the smaller side-chain of glycine (as compared to arginine in the wild-type enzyme) avoids a key clash with the acyl group of the N-acyl homoserine lactone substrate.
Conclusions: Mutation of a single amino acid residue successfully converted a GCA (with no detectable activity against AHLs) into an AHL acylase. This approach may be useful for further engineering of 'quorum quenching' enzymes.
Keywords: Glutaryl-7-aminocephalosporanic acid acylase; N-acyl-homoserine lactone acylase; Protein engineering; Quorum quenching; Site‐saturation mutagenesis.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

