In silico approach for identifying natural lead molecules against SARS-COV-2
- PMID: 33892297
- PMCID: PMC8042570
- DOI: 10.1016/j.jmgm.2021.107916
In silico approach for identifying natural lead molecules against SARS-COV-2
Abstract
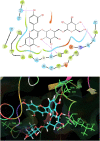
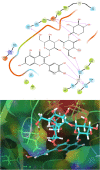
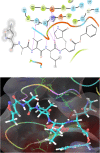
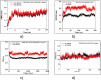
The life challenging COVID-19 disease caused by the SARS-CoV-2 virus has greatly impacted smooth survival worldwide since its discovery in December 2019. Currently, it is one of the major threats to humanity. Moreover, any specific drug or vaccine unavailability against COVID-19 forces to discover a new drug on an urgent basis. Viral cycle inhibition could be one possible way to prevent the further genesis of this viral disease, which can be contributed by drug repurposing techniques or screening of small bioactive natural molecules against already validated targets of COVID-19. The main protease (Mpro) responsible for producing functional proteins from polyprotein is an important key step for SARS-CoV-2 virion replication. Natural product or herbal based formulations are an important platform for potential therapeutics and lead compounds in the drug discovery process. Therefore, here we have screened >53,500 bioactive natural molecules from six different natural product databases against Mpro (PDB ID: 6LU7) of COVID-19 through computational study. Further, the top three molecules were subjected to pharmacokinetics evaluation, which is an important factor that reduces the drug failure rate. Moreover, the top three screened molecules (C00014803, C00006660, ANLT0001) were further validated by a molecular dynamics study under a condition similar to the physiological one. Relative binding energy analysis of three lead molecules indicated that C00014803 possess highest binding affinity among all three hits. These extensive studies can be a significant foundation for developing a therapeutic agent against COVID-19 through vet lab studies.
Keywords: COVID-19; In silico; Main protease; Molecular dynamics; Natural products; Pharmacokinetic parameters.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- WHO Weekly epidemiological update on COVID-19 – 16 March 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update---...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

