Drosophila primary microRNA-8 encodes a microRNA-encoded peptide acting in parallel of miR-8
- PMID: 33892772
- PMCID: PMC8063413
- DOI: 10.1186/s13059-021-02345-8
Drosophila primary microRNA-8 encodes a microRNA-encoded peptide acting in parallel of miR-8
Abstract
Background: Recent genome-wide studies of many species reveal the existence of a myriad of RNAs differing in size, coding potential and function. Among these are the long non-coding RNAs, some of them producing functional small peptides via the translation of short ORFs. It now appears that any kind of RNA presumably has a potential to encode small peptides. Accordingly, our team recently discovered that plant primary transcripts of microRNAs (pri-miRs) produce small regulatory peptides (miPEPs) involved in auto-regulatory feedback loops enhancing their cognate microRNA expression which in turn controls plant development. Here we investigate whether this regulatory feedback loop is present in Drosophila melanogaster.
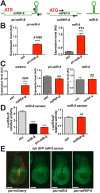
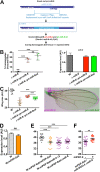
Results: We perform a survey of ribosome profiling data and reveal that many pri-miRNAs exhibit ribosome translation marks. Focusing on miR-8, we show that pri-miR-8 can produce a miPEP-8. Functional assays performed in Drosophila reveal that miPEP-8 affects development when overexpressed or knocked down. Combining genetic and molecular approaches as well as genome-wide transcriptomic analyses, we show that miR-8 expression is independent of miPEP-8 activity and that miPEP-8 acts in parallel to miR-8 to regulate the expression of hundreds of genes.
Conclusion: Taken together, these results reveal that several Drosophila pri-miRs exhibit translation potential. Contrasting with the mechanism described in plants, these data shed light on the function of yet undescribed primary-microRNA-encoded peptides in Drosophila and their regulatory potential on genome expression.
Keywords: Drosophila; Small peptides; lncRNA; miPEP; miR-8; sORF.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

