Finding associations in a heterogeneous setting: statistical test for aberration enrichment
- PMID: 33892787
- PMCID: PMC8066476
- DOI: 10.1186/s13073-021-00864-4
Finding associations in a heterogeneous setting: statistical test for aberration enrichment
Abstract
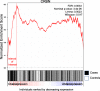
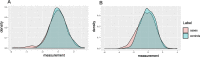
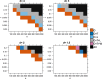
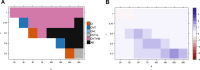
Most two-group statistical tests find broad patterns such as overall shifts in mean, median, or variance. These tests may not have enough power to detect effects in a small subset of samples, e.g., a drug that works well only on a few patients. We developed a novel statistical test targeting such effects relevant for clinical trials, biomarker discovery, feature selection, etc. We focused on finding meaningful associations in complex genetic diseases in gene expression, miRNA expression, and DNA methylation. Our test outperforms traditional statistical tests in simulated and experimental data and detects potentially disease-relevant genes with heterogeneous effects.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

