Interstitial Deletions Generating Fusion Genes
- PMID: 33893073
- PMCID: PMC8126330
- DOI: 10.21873/cgp.20251
Interstitial Deletions Generating Fusion Genes
Abstract
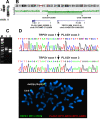
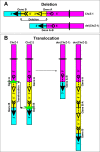
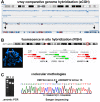
A fusion gene is the physical juxtaposition of two different genes resulting in a structure consisting of the head of one gene and the tail of the other. Gene fusion is often a primary neoplasia-inducing event in leukemias, lymphomas, solid malignancies as well as benign tumors. Knowledge about fusion genes is crucial not only for our understanding of tumorigenesis, but also for the diagnosis, prognostication, and treatment of cancer. Balanced chromosomal rearrangements, in particular translocations and inversions, are the most frequent genetic events leading to the generation of fusion genes. In the present review, we summarize the existing knowledge on chromosome deletions as a mechanism for fusion gene formation. Such deletions are mostly submicroscopic and, hence, not detected by cytogenetic analyses but by array comparative genome hybridization (aCGH) and/or high throughput sequencing (HTS). They are found across the genome in a variety of neoplasias. As tumors are increasingly analyzed using aCGH and HTS, it is likely that more interstitial deletions giving rise to fusion genes will be found, significantly impacting our understanding and treatment of cancer.
Keywords: Interstitial deletion; chromosome; cytogenetics; fusion gene; review.
Copyright© 2021, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
The Authors declare that they have no potential conflicts of interest with regards to this study.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
