Clinical Utility of Functional RNA Analysis for the Reclassification of Splicing Gene Variants in Hereditary Cancer
- PMID: 33893081
- PMCID: PMC8126331
- DOI: 10.21873/cgp.20259
Clinical Utility of Functional RNA Analysis for the Reclassification of Splicing Gene Variants in Hereditary Cancer
Abstract
Background: Classification of splicing variants (SVs) in genes associated with hereditary cancer is often challenging. The aim of this study was to investigate the occurrence of SVs in hereditary cancer genes and the clinical utility of RNA analysis.
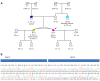
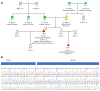
Material and methods: 1518 individuals were tested for cancer predisposition, using a Next Generation Sequencing (NGS) panel of 36 genes. Splicing variant analysis was performed using RT-PCR and Sanger Sequencing.
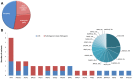
Results: In total, 34 different SVs were identified, 53% of which were classified as pathogenic or likely pathogenic. The remaining 16 variants were initially classified as Variant of Uncertain Significance (VUS). RNA analysis was performed for 3 novel variants.
Conclusion: The RNA analysis assisted in the reclassification of 20% of splicing variants from VUS to pathogenic. RNA analysis is essential in the case of uncharacterized splicing variants, for proper classification and personalized management of these patients.
Keywords: NGS; RNA analysis; Splicing variant; cancer genes.
Copyright© 2021, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
There are no conflicts to declare.
Figures

References
-
- Castéra L, Krieger S, Rousselin A, Legros A, Baumann JJ, Bruet O, Brault B, Fouillet R, Goardon N, Letac O, Baert-Desurmont S, Tinat J, Bera O, Dugast C, Berthet P, Polycarpe F, Layet V, Hardouin A, Frébourg T, Vaur D. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet. 2014;22(11):1305–1313. doi: 10.1038/ejhg.2014.16. - DOI - PMC - PubMed
-
- Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, Timms K, Garber JE, Herold C, Ellisen L, Krejdovsky J, DeLeonardis K, Sedgwick K, Soltis K, Roa B, Wenstrup RJ, Hartman AR. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121(1):25–33. doi: 10.1002/cncr.29010. - DOI - PubMed
-
- Susswein LR, Marshall ML, Nusbaum R, Vogel Postula KJ, Weissman SM, Yackowski L, Vaccari EM, Bissonnette J, Booker JK, Cremona ML, Gibellini F, Murphy PD, Pineda-Alvarez DE, Pollevick GD, Xu Z, Richard G, Bale S, Klein RT, Hruska KS, Chung WK. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med. 2016;18(8):823–832. doi: 10.1038/gim.2015.166. - DOI - PMC - PubMed
-
- Tsaousis GN, Papadopoulou E, Apessos A, Agiannitopoulos K, Pepe G, Kampouri S, Diamantopoulos N, Floros T, Iosifidou R, Katopodi O, Koumarianou A, Markopoulos C, Papazisis K, Venizelos V, Xanthakis I, Xepapadakis G, Banu E, Eniu DT, Negru S, Stanculeanu DL, Ungureanu A, Ozmen V, Tansan S, Tekinel M, Yalcin S, Nasioulas G. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer. 2019;19(1):535. doi: 10.1186/s12885-019-5756-4. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
