The structural plasticity of nucleic acid duplexes revealed by WAXS and MD
- PMID: 33893104
- PMCID: PMC8064643
- DOI: 10.1126/sciadv.abf6106
The structural plasticity of nucleic acid duplexes revealed by WAXS and MD
Abstract
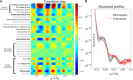
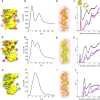
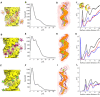
Double-stranded DNA (dsDNA) and RNA (dsRNA) helices display an unusual structural diversity. Some structural variations are linked to sequence and may serve as signaling units for protein-binding partners. Therefore, elucidating the mechanisms and factors that modulate these variations is of fundamental importance. While the structural diversity of dsDNA has been extensively studied, similar studies have not been performed for dsRNA. Because of the increasing awareness of RNA's diverse biological roles, such studies are timely and increasingly important. We integrate solution x-ray scattering at wide angles (WAXS) with all-atom molecular dynamics simulations to explore the conformational ensemble of duplex topologies for different sequences and salt conditions. These tightly coordinated studies identify robust correlations between features in the WAXS profiles and duplex geometry and enable atomic-level insights into the structural diversity of DNA and RNA duplexes. Notably, dsRNA displays a marked sensitivity to the valence and identity of its associated cations.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures






References
-
- Conn G. L., Draper D. E., Lattman E. E., Gittis A. G., Crystal structure of a conserved ribosomal protein-RNA complex. Science 284, 1171–1174 (1999). - PubMed
-
- Bukhman Y. V., Draper D. E., Affinities and selectivities of divalent cation binding sites within an RNA tertiary structure. J. Mol. Biol. 273, 1020–1031 (1997). - PubMed
-
- Lu X.-J., Shakked Z., Olson W. K., A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 300, 819–840 (2000). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

