Probiotic Bacillus Affects Enterococcus faecalis Antibiotic Resistance Transfer by Interfering with Pheromone Signaling Cascades
- PMID: 33893118
- PMCID: PMC8316027
- DOI: 10.1128/AEM.00442-21
Probiotic Bacillus Affects Enterococcus faecalis Antibiotic Resistance Transfer by Interfering with Pheromone Signaling Cascades
Abstract
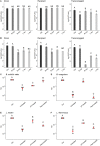
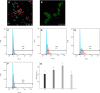
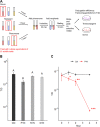
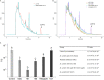
Enterococcus faecalis, a member of the commensal flora in the human gastrointestinal tract, has become a threatening nosocomial pathogen because it has developed resistance to many known antibiotics. More concerningly, resistance gene-carrying E. faecalis cells may transfer antibiotic resistance to resistance-free E. faecalis cells through their unique quorum sensing-mediated plasmid transfer system. Therefore, we investigated the role of probiotic bacteria in the transfer frequency of the antibiotic resistance plasmid pCF10 in E. faecalis populations to mitigate the spread of antibiotic resistance. Bacillus subtilis subsp. natto is a probiotic strain isolated from Japanese fermented soybean foods, and its culture fluid potently inhibited pCF10 transfer by suppressing peptide pheromone activity from chromosomally encoded CF10 (cCF10) without inhibiting E. faecalis growth. The inhibitory effect was attributed to at least one 30- to 50-kDa extracellular protease present in B. subtilis subsp. natto. Nattokinase of B. subtilis subsp. natto was involved in the inhibition of pCF10 transfer and cleaved cCF10 (LVTLVFV) into LVTL plus VFV fragments. Moreover, the cleavage product LVTL (L peptide) interfered with the conjugative transfer of pCF10. In addition to cCF10, faecalis-cAM373 and gordonii-cAM373, which are mating inducers of vancomycin-resistant E. faecalis, were also cleaved by nattokinase, indicating that B. subtilis subsp. natto can likely interfere with vancomycin resistance transfer in E. faecalis. Our work shows the feasibility of applying fermentation products of B. subtilis subsp. natto and L peptide to mitigate E. faecalis antibiotic resistance transfer. IMPORTANCE Enterococcus faecalis is considered a leading cause of hospital-acquired infections. Treatment of these infections has become a major challenge for clinicians because some E. faecalis strains are resistant to multiple clinically used antibiotics. Moreover, antibiotic resistance genes can undergo efficient intra- and interspecies transfer via E. faecalis peptide pheromone-mediated plasmid transfer systems. Therefore, this study provided the first experimental demonstration that probiotics are a feasible approach for interfering with conjugative plasmid transfer between E. faecalis strains to stop the transfer of antibiotic resistance. We found that the extracellular protease(s) of Bacillus subtilis subsp. natto cleaved peptide pheromones without affecting the growth of E. faecalis, thereby reducing the frequency of conjugative plasmid transfer. In addition, a specific cleaved pheromone fragment interfered with conjugative plasmid transfer. These findings provide a potential probiotic-based method for interfering with the transfer of antibiotic resistance between E. faecalis strains.
Keywords: Bacillus subtilis subsp. natto; Enterococcus faecalis; antibiotic resistance; nattokinase; pheromone-inducible conjugative plasmid transfer; probiotics.
Figures


Similar articles
-
Enterococcus faecalis Sex Pheromone cCF10 Enhances Conjugative Plasmid Transfer In Vivo.mBio. 2018 Feb 13;9(1):e00037-18. doi: 10.1128/mBio.00037-18. mBio. 2018. PMID: 29440568 Free PMC article.
-
Isolation of VanB-type Enterococcus faecalis strains from nosocomial infections: first report of the isolation and identification of the pheromone-responsive plasmids pMG2200, Encoding VanB-type vancomycin resistance and a Bac41-type bacteriocin, and pMG2201, encoding erythromycin resistance and cytolysin (Hly/Bac).Antimicrob Agents Chemother. 2009 Feb;53(2):735-47. doi: 10.1128/AAC.00754-08. Epub 2008 Nov 24. Antimicrob Agents Chemother. 2009. PMID: 19029325 Free PMC article.
-
ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF.J Bacteriol. 2002 Feb;184(4):1155-62. doi: 10.1128/jb.184.4.1155-1162.2002. J Bacteriol. 2002. PMID: 11807076 Free PMC article.
-
The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution.Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1185-93. doi: 10.1098/rstb.2007.2043. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360276 Free PMC article. Review.
-
Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response.Peptides. 2001 Oct;22(10):1529-39. doi: 10.1016/s0196-9781(01)00489-2. Peptides. 2001. PMID: 11587782 Review.
Cited by
-
Translating eco-evolutionary biology into therapy to tackle antibiotic resistance.Nat Rev Microbiol. 2023 Oct;21(10):671-685. doi: 10.1038/s41579-023-00902-5. Epub 2023 May 19. Nat Rev Microbiol. 2023. PMID: 37208461 Review.
-
Bacillus subtilis natto Derivatives Inhibit Enterococcal Biofilm Formation via Restructuring of the Cell Envelope.Front Microbiol. 2021 Dec 9;12:785351. doi: 10.3389/fmicb.2021.785351. eCollection 2021. Front Microbiol. 2021. PMID: 34956152 Free PMC article.
-
Enterococcus faecalis: implications for host health.World J Microbiol Biotechnol. 2024 May 4;40(6):190. doi: 10.1007/s11274-024-04007-w. World J Microbiol Biotechnol. 2024. PMID: 38702495 Review.
-
The Networked Interaction between Probiotics and Intestine in Health and Disease: A Promising Success Story.Microorganisms. 2024 Jan 18;12(1):194. doi: 10.3390/microorganisms12010194. Microorganisms. 2024. PMID: 38258020 Free PMC article. Review.
-
What are the missing pieces needed to stop antibiotic resistance?Microb Biotechnol. 2023 Oct;16(10):1900-1923. doi: 10.1111/1751-7915.14310. Epub 2023 Jul 7. Microb Biotechnol. 2023. PMID: 37417823 Free PMC article. Review.
References
-
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

