Cdk5 and GSK3β inhibit fast endophilin-mediated endocytosis
- PMID: 33893293
- PMCID: PMC8065113
- DOI: 10.1038/s41467-021-22603-4
Cdk5 and GSK3β inhibit fast endophilin-mediated endocytosis
Abstract
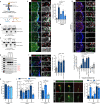
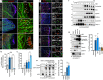
Endocytosis mediates the cellular uptake of micronutrients and cell surface proteins. Fast Endophilin-mediated endocytosis, FEME, is not constitutively active but triggered upon receptor activation. High levels of growth factors induce spontaneous FEME, which can be suppressed upon serum starvation. This suggested a role for protein kinases in this growth factor receptor-mediated regulation. Using chemical and genetic inhibition, we find that Cdk5 and GSK3β are negative regulators of FEME. They antagonize the binding of Endophilin to Dynamin-1 and to CRMP4, a Plexin A1 adaptor. This control is required for proper axon elongation, branching and growth cone formation in hippocampal neurons. The kinases also block the recruitment of Dynein onto FEME carriers by Bin1. As GSK3β binds to Endophilin, it imposes a local regulation of FEME. Thus, Cdk5 and GSK3β are key regulators of FEME, licensing cells for rapid uptake by the pathway only when their activity is low.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- BB/R01551X/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104913/Z/14/ZBM/WT_/Wellcome Trust/United Kingdom
- BB/R01552X/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/N025644/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

