Introduction, Transmission Dynamics, and Fate of Early Severe Acute Respiratory Syndrome Coronavirus 2 Lineages in Santa Clara County, California
- PMID: 33893501
- PMCID: PMC8135363
- DOI: 10.1093/infdis/jiab199
Introduction, Transmission Dynamics, and Fate of Early Severe Acute Respiratory Syndrome Coronavirus 2 Lineages in Santa Clara County, California
Abstract
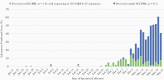
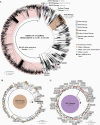
We combined viral genome sequencing with contact tracing to investigate introduction and evolution of severe acute respiratory syndrome coronavirus 2 lineages in Santa Clara County, California, from 27 January to 21 March 2020. From 558 persons with coronavirus disease 2019, 101 genomes from 143 available clinical samples comprised 17 lineages, including SCC1 (n = 41), WA1 (n = 9; including the first 2 reported deaths in the United States, with postmortem diagnosis), D614G (n = 4), ancestral Wuhan Hu-1 (n = 21), and 13 others (n = 26). Public health intervention may have curtailed the persistence of lineages that appeared transiently during February and March. By August, only D614G lineages introduced after 21 March were circulating in Santa Clara County.
Keywords: COVID-19; D614G lineage; SARS-CoV-2; epidemiology; public health surveillance; viral evolution; viral whole-genome sequencing.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

