Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly
- PMID: 33893684
- PMCID: PMC8360199
- DOI: 10.1111/tpj.15289
Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly
Abstract
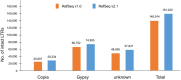
Until recently, achieving a reference-quality genome sequence for bread wheat was long thought beyond the limits of genome sequencing and assembly technology, primarily due to the large genome size and > 80% repetitive sequence content. The release of the chromosome scale 14.5-Gb IWGSC RefSeq v1.0 genome sequence of bread wheat cv. Chinese Spring (CS) was, therefore, a milestone. Here, we used a direct label and stain (DLS) optical map of the CS genome together with a prior nick, label, repair and stain (NLRS) optical map, and sequence contigs assembled with Pacific Biosciences long reads, to refine the v1.0 assembly. Inconsistencies between the sequence and maps were reconciled and gaps were closed. Gap filling and anchoring of 279 unplaced scaffolds increased the total length of pseudomolecules by 168 Mb (excluding Ns). Positions and orientations were corrected for 233 and 354 scaffolds, respectively, representing 10% of the genome sequence. The accuracy of the remaining 90% of the assembly was validated. As a result of the increased contiguity, the numbers of transposable elements (TEs) and intact TEs have increased in IWGSC RefSeq v2.1 compared with v1.0. In total, 98% of the gene models identified in v1.0 were mapped onto this new assembly through development of a dedicated approach implemented in the MAGAAT pipeline. The numbers of high-confidence genes on pseudomolecules have increased from 105 319 to 105 534. The reconciled assembly enhances the utility of the sequence for genetic mapping, comparative genomics, gene annotation and isolation, and more general studies on the biology of wheat.
Keywords: Hi-C; direct label and stain; gene collinearity; pseudomolecule; transposable element.
© 2021 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures
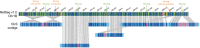
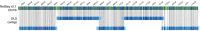

References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. - PubMed
-
- Avni, R., Nave, M., Eilam, T., Sela, H., Alekperov, C., Peleg, Z.et al. (2014) Ultra‐dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Molecular Breeding, 34, 1549–1562.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

