Multicolor single-molecule FRET for DNA and RNA processes
- PMID: 33894656
- PMCID: PMC8528908
- DOI: 10.1016/j.sbi.2021.03.005
Multicolor single-molecule FRET for DNA and RNA processes
Abstract
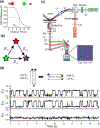
Single-molecule fluorescence resonance energy transfer (smFRET) is a useful tool for observing the dynamics of protein-nucleic acid interactions. Although most smFRET measurements have used two fluorophores, multicolor smFRET measurements using more than two fluorophores offer more information about how protein-nucleic acid complexes dynamically move, assemble, and disassemble. Multicolor smFRET experiments include three or more fluorophores and at least one donor-acceptor pair. This review highlights how multicolor smFRET is being used to probe the dynamics of three different classes of biochemical processes-protein-DNA interactions, chromatin remodeling, and protein translation.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest statement Nothing declared.
Figures



References
-
- Abbe E: Beiträge zur Theorie des Mikroskops und der mikrosk opischen Wahrnehmung: I. Die Construction von Mikroskopen auf Grund der Theorie. Arch für mikroskopische Anat 1873, 9:413–418.
-
- Hell SW, Wichmann J: Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 1994, 19:780. - PubMed
-
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF: Imaging intracellular fluorescent proteins at nanometer resolution. Science (80- ) 2006, 313:1642–1645. - PubMed
-
- Förster T: Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann Phys 1948, 437:55–75.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

