Proteomic profiling of soft tissue sarcomas with SWATH mass spectrometry
- PMID: 33895336
- PMCID: PMC8135130
- DOI: 10.1016/j.jprot.2021.104236
Proteomic profiling of soft tissue sarcomas with SWATH mass spectrometry
Abstract
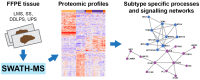
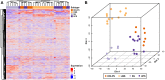
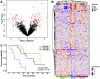
Soft tissue sarcomas (STS) are a group of rare and heterogeneous cancers. While large-scale genomic and epigenomic profiling of STS have been undertaken, proteomic analysis has thus far been limited. Here we utilise sequential window acquisition of all theoretical fragment ion spectra mass spectrometry (SWATH-MS) for proteomic profiling of formalin fixed paraffin embedded (FFPE) specimens from a cohort of STS patients (n = 36) across four histological subtypes (leiomyosarcoma, synovial sarcoma, undifferentiated pleomorphic sarcoma and dedifferentiated liposarcoma). We quantified 2951 proteins across all cases and show that there is a significant enrichment of gene sets associated with smooth muscle contraction in leiomyosarcoma, RNA splicing regulation in synovial sarcoma and leukocyte activation in undifferentiated pleomorphic sarcoma. We further identified a subgroup of STS cases that have a distinct expression profile in a panel of proteins, with worse survival outcomes when compared to the rest of the cohort. Our study highlights the value of comprehensive proteomic characterisation as a means to identify histotype-specific STS profiles that describe key biological pathways of clinical and therapeutic relevance; as well as for discovering new prognostic biomarkers in this group of rare and difficult-to-treat diseases.
Keywords: Biomarkers; FFPE; Mass spectrometry; Proteomics; SWATH MS; Soft tissue sarcoma.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- WHO . 5th ed. World Health Organization; 2020. Classification of Tumours Editorial Board, Soft Tissue and Bone Tumours.
-
- Chudasama P., Mughal S.S., Sanders M.A., Hubschmann D., Chung I., Deeg K.I., Wong S.H., Rabe S., Hlevnjak M., Zapatka M., Ernst A., Kleinheinz K., Schlesner M., Sieverling L., Klink B., Schrock E., Hoogenboezem R.M., Kasper B., Heilig C.E., Egerer G., Wolf S., von Kalle C., Eils R., Stenzinger A., Weichert W., Glimm H., Groschel S., Kopp H.G., Omlor G., Lehner B., Bauer S., Schimmack S., Ulrich A., Mechtersheimer G., Rippe K., Brors B., Hutter B., Renner M., Hohenberger P., Scholl C., Frohling S. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat. Commun. 2018;9(1):144. - PMC - PubMed
-
- Le Guellec S., Lesluyes T., Sarot E., Valle C., Filleron T., Rochaix P., Valentin T., Perot G., Coindre J.M., Chibon F. Validation of the Complexity INdex in SARComas prognostic signature on formalin-fixed, paraffin-embedded, soft-tissue sarcomas. Ann. Oncol. 2018;29(8):1828–1835. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

