An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19
- PMID: 33895786
- PMCID: PMC8065335
- DOI: 10.1038/s41392-021-00568-6
An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19
Abstract
The global spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) requires an urgent need to find effective therapeutics for the treatment of coronavirus disease 2019 (COVID-19). In this study, we developed an integrative drug repositioning framework, which fully takes advantage of machine learning and statistical analysis approaches to systematically integrate and mine large-scale knowledge graph, literature and transcriptome data to discover the potential drug candidates against SARS-CoV-2. Our in silico screening followed by wet-lab validation indicated that a poly-ADP-ribose polymerase 1 (PARP1) inhibitor, CVL218, currently in Phase I clinical trial, may be repurposed to treat COVID-19. Our in vitro assays revealed that CVL218 can exhibit effective inhibitory activity against SARS-CoV-2 replication without obvious cytopathic effect. In addition, we showed that CVL218 can interact with the nucleocapsid (N) protein of SARS-CoV-2 and is able to suppress the LPS-induced production of several inflammatory cytokines that are highly relevant to the prevention of immunopathology induced by SARS-CoV-2 infection.
Conflict of interest statement
J.Z. is founder of Silexon AI Technology Co. Ltd. and has an equity interest. X.S. is founder and CEO of Convalife (Shanghai) Co. Ltd. and has an equity interest.
Figures
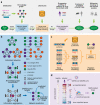
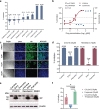
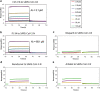
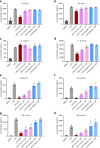
References
-
- Hong L, Lin J, Li S, et al. A novel machine learning framework for automated biomedical relation extraction from large-scale literature repositories. Nat. Mach. Intell. 2020;2:347–355. doi: 10.1038/s42256-020-0189-y. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

