Mutation Rates and Selection on Synonymous Mutations in SARS-CoV-2
- PMID: 33895815
- PMCID: PMC8135539
- DOI: 10.1093/gbe/evab087
Mutation Rates and Selection on Synonymous Mutations in SARS-CoV-2
Abstract
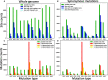
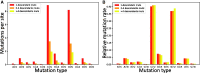
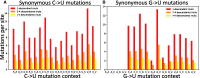
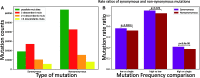
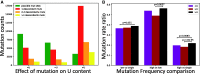
The COVID-19 pandemic has seen an unprecedented response from the sequencing community. Leveraging the sequence data from more than 140,000 SARS-CoV-2 genomes, we study mutation rates and selective pressures affecting the virus. Understanding the processes and effects of mutation and selection has profound implications for the study of viral evolution, for vaccine design, and for the tracking of viral spread. We highlight and address some common genome sequence analysis pitfalls that can lead to inaccurate inference of mutation rates and selection, such as ignoring skews in the genetic code, not accounting for recurrent mutations, and assuming evolutionary equilibrium. We find that two particular mutation rates, G →U and C →U, are similarly elevated and considerably higher than all other mutation rates, causing the majority of mutations in the SARS-CoV-2 genome, and are possibly the result of APOBEC and ROS activity. These mutations also tend to occur many times at the same genome positions along the global SARS-CoV-2 phylogeny (i.e., they are very homoplasic). We observe an effect of genomic context on mutation rates, but the effect of the context is overall limited. Although previous studies have suggested selection acting to decrease U content at synonymous sites, we bring forward evidence suggesting the opposite.
Keywords: COVID-19; SARS-CoV-2; mutation; selection; sequencing; viral genomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Update of
-
Mutation rates and selection on synonymous mutations in SARS-CoV-2.bioRxiv [Preprint]. 2021 Jan 14:2021.01.14.426705. doi: 10.1101/2021.01.14.426705. bioRxiv. 2021. Update in: Genome Biol Evol. 2021 May 7;13(5):evab087. doi: 10.1093/gbe/evab087. PMID: 33469589 Free PMC article. Updated. Preprint.
References
-
- Clemente F, Vogl C.. 2012. Evidence for complex selection on four-fold degenerate sites in drosophila melanogaster. J Evol Biol. 25(12):2582–2595. - PubMed
-
- Cuevas JM, Domingo-Calap P, Sanjuán R.. 2012. The fitness effects of synonymous mutations in DNA and RNA viruses. Mol Biol Evol. 29(1):17–20. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

