Endosperm-based incompatibilities in hybrid monkeyflowers
- PMID: 33895820
- PMCID: PMC8364248
- DOI: 10.1093/plcell/koab117
Endosperm-based incompatibilities in hybrid monkeyflowers
Erratum in
-
Erratum to: Endosperm-based incompatibilities in hybrid monkeyflowers.Plant Cell. 2022 Mar 29;34(4):1418-1419. doi: 10.1093/plcell/koab259. Plant Cell. 2022. PMID: 34850230 Free PMC article. No abstract available.
Abstract
Endosperm is an angiosperm innovation central to their reproduction whose development, and thus seed viability, is controlled by genomic imprinting, where expression from certain genes is parent-specific. Unsuccessful imprinting has been linked to failed inter-specific and inter-ploidy hybridization. Despite their importance in plant speciation, the underlying mechanisms behind these endosperm-based barriers remain poorly understood. Here, we describe one such barrier between diploid Mimulus guttatus and tetraploid Mimulus luteus. The two parents differ in endosperm DNA methylation, expression dynamics, and imprinted genes. Hybrid seeds suffer from underdeveloped endosperm, reducing viability, or arrested endosperm and seed abortion when M. guttatus or M. luteus is seed parent, respectively, and transgressive methylation and expression patterns emerge. The two inherited M. luteus subgenomes, genetically distinct but epigenetically similar, are expressionally dominant over the M. guttatus genome in hybrid embryos and especially their endosperm, where paternal imprints are perturbed. In aborted seeds, de novo methylation is inhibited, potentially owing to incompatible paternal instructions of imbalanced dosage from M. guttatus imprints. We suggest that diverged epigenetic/regulatory landscapes between parental genomes induce epigenetic repatterning and global shifts in expression, which, in endosperm, may uniquely facilitate incompatible interactions between divergent imprinting schemes, potentially driving rapid barriers.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
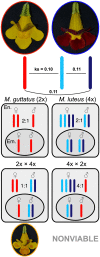





Comment in
-
Genome shock in monkeyflower hybrids.Plant Cell. 2021 Aug 13;33(7):2097-2098. doi: 10.1093/plcell/koab118. Plant Cell. 2021. PMID: 35233602 Free PMC article. No abstract available.
References
-
- Alger EI, Edger PP (2020) One subgenome to rule them all: underlying mechanisms of subgenome dominance. Curr Opin Plant Biol 54: 108–113 - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

